5GZ8
 
 | |
1MT8
 
 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | ACETATE ION, Capsid-p2 substrate peptide of HIV-1 Gag polyprotein, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
5UUC
 
 | | Tetragonal thermolysin cryocooled to 100 K with 50% mpd as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60000932 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5GUW
 
 | | Complex of Cytochrome cd1 Nitrite Reductase and Nitric Oxide Reductase in Denitrification of Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Terasaka, E, Sugimoto, H, Shiro, Y, Tosha, T. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dynamics of nitric oxide controlled by protein complex in bacterial system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1EBG
 
 | |
6L99
 
 | | Zinc finger of RING finger protein 144A | | Descriptor: | E3 ubiquitin-protein ligase RNF144A, ZINC ION | | Authors: | Miyamoto, K. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc finger of RING finger protein 144A
To Be Published
|
|
6D5S
 
 | | Hexagonal thermolysin cryocooled to 100 K with 50% MPD as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, LYSINE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00003552 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
9COR
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | (2R)-3-phenyl-2-{[(2S)-3-phenyl-2-{[N-(trifluoroacetyl)glycyl]amino}propyl]sulfanyl}-N-[2-(pyridin-3-yl)ethyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9COS
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | (2R)-3-phenyl-2-{[(2S)-3-phenyl-2-{[N-(trifluoroacetyl)glycyl]amino}propyl]sulfanyl}-N-[3-(pyridin-3-yl)propyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9COX
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, N-[(2S)-1-(naphthalen-1-yl)-3-{[(2S)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}propan-2-yl]pyridine-3-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9COV
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, N-[(2S)-1-(1H-indol-3-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}propan-2-yl]pyridine-3-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9COW
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, GLYCEROL, N-[(2S)-1-(naphthalen-1-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}propan-2-yl]pyridine-3-carboxamide, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9COY
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, N-[(2S)-1-([1,1'-biphenyl]-4-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[3-(pyridin-3-yl)propyl]amino}propan-2-yl]sulfanyl}propan-2-yl]-2,2-dimethylpropanamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9COT
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | (2R)-3-phenyl-2-{[(2S)-3-phenyl-2-(2,2,2-trifluoroacetamido)propyl]sulfanyl}-N-[3-(pyridin-3-yl)propyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
9COU
 
 | | Crystal structure of CYP3A4 bound to an inhibitor | | Descriptor: | (2R)-2-{[(2S)-2-(benzenesulfonamido)-3-phenylpropyl]sulfanyl}-3-phenyl-N-[3-(pyridin-3-yl)propyl]propanamide, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioiukova, I.F. | | Deposit date: | 2024-07-17 | | Release date: | 2025-01-08 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Evaluation of Larger Side-Group Functionalities and the Side/End-Group Interplay in Ritonavir-Like Inhibitors of CYP3A4.
Chem.Biol.Drug Des., 105, 2025
|
|
6D5R
 
 | |
6D5Q
 
 | |
1N49
 
 | | Viability of a Drug-Resistant HIV-1 Protease Variant: Structural Insights for Better Anti-Viral Therapy | | Descriptor: | Protease, RITONAVIR | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Viability of a Drug-Resistant Human Immunodeficiency Virus Type 1 Protease Variant:
Structural Insights for Better Antiviral Therapy
J.VIROL., 77, 2003
|
|
6D5N
 
 | | Hexagonal thermolysin (295) in the presence of 50% xylose | | Descriptor: | CALCIUM ION, LYSINE, Thermolysin, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00003672 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D5U
 
 | |
5UN3
 
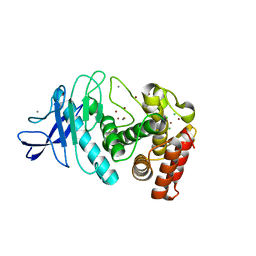 | | Tetragonal thermolysin (295 K) in the presence of 50% xylose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermolysin, ... | | Authors: | Juers, D. | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60004437 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5UUB
 
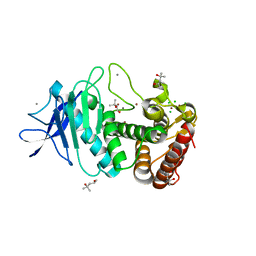 | |
1WTG
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-biphenylalanine-Gln-p-aminobenzamidine | | Descriptor: | 2-(3-BIPHENYL-4-YL-2-ETHANESULFONYLAMINO-PROPIONYLAMINO)-PENTANEDIOIC ACID 5-AMIDE 1-(4-CARBAMIMIDOYL-BENZYLAMIDE), CALCIUM ION, Coagulation factor VII, ... | | Authors: | Kadono, S, Sakamoto, S, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Kitazawa, K, Yoshihashi, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Kodama, M, Haramura, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-11-23 | | Release date: | 2005-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel interactions of large P3 moiety and small P4 moiety in the binding of the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 326, 2005
|
|
2ALG
 
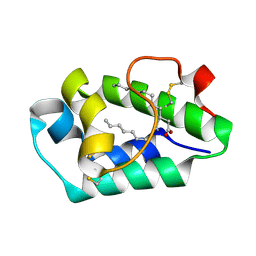 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
6DR5
 
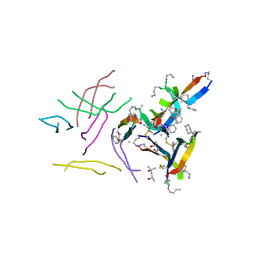 | |
