4TK5
 
 | | Crystal Structure of human Tankyrase 2 in complex with EB47. | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7ND7
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
5MIY
 
 | | Crystal structure of the E3 ubiquitin ligase RavN from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin ligase RavN, SODIUM ION, ... | | Authors: | Lucas, M, Abascal-Palacios, G, Rojas, A.L, Hierro, A. | | Deposit date: | 2016-11-29 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | RavN is a member of a previously unrecognized group of Legionella pneumophila E3 ubiquitin ligases.
PLoS Pathog., 14, 2018
|
|
7OX3
 
 | | Fab 6D3: hIL-9 complex | | Descriptor: | Heavy chain (Fab 6D3), Interleukin-9, Light chain (Fab 6D3), ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
4TM5
 
 | |
7OX2
 
 | | Fab 6E2: hIL-9 complex | | Descriptor: | Heavy chain (Fab 6E2), Interleukin-9, Light chain (Fab 6E2), ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
6J11
 
 | | MERS-CoV spike N-terminal domain and 7D10 scFv complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-terminal domain of Spike glycoprotein, ... | | Authors: | Zhou, H, Zhang, S, Zhang, S, Tang, W, Wang, X. | | Deposit date: | 2018-12-27 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein.
Nat Commun, 10, 2019
|
|
7OX1
 
 | | Fab 7D6: hIL-9 complex | | Descriptor: | Heavy chain (Fab 7D6), Interleukin-9, Light chain (Fab 7D6) | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
4TN2
 
 | | NS5b in complex with lactam-thiophene carboxylic acids | | Descriptor: | 3-[(2R)-2-cyclohexyl-5-oxopyrrolidin-1-yl]-5-phenylthiophene-2-carboxylic acid, Genome polyprotein | | Authors: | Chopra, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and synthesis of lactam-thiophene carboxylic acids as potent hepatitis C virus polymerase inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7ND8
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-384 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-384 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7OX4
 
 | | Mouse interleukin-9 in complex with Fab 35D8. | | Descriptor: | ACETATE ION, Heavy chain (Fab 35D8), Interleukin-9, ... | | Authors: | De Vos, T, Savvides, S.N. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
7NDC
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
4TMD
 
 | | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis | | Descriptor: | IODIDE ION, Uncharacterized protein | | Authors: | Horanyi, P.S, Dranow, D.M, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-06-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Putative uncharacterized protein (Rv0999 ortholog) from Mycobacterium smegmatis
To Be Published
|
|
4TN4
 
 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with glycine and a novel pyrazolopyran 33G: (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Witschel, M.C. | | Deposit date: | 2014-06-03 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT): Cocrystal Structures of Pyrazolopyrans with Potent Blood- and Liver-Stage Activities.
J.Med.Chem., 58, 2015
|
|
7OYN
 
 | | Carbonic anhydrase II in complex with Hit3 (MH57) | | Descriptor: | Carbonic anhydrase 2, Hit3 (MH57), ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2021-06-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
6X7H
 
 | |
4TMP
 
 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
7OYR
 
 | | Carbonic anhydrase II in complex with Hit3-t4 (MH181) | | Descriptor: | Carbonic anhydrase 2, Hit3-t4 (MH181), ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2021-06-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
7NDD
 
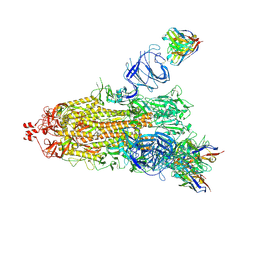 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
4TN0
 
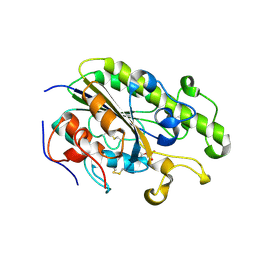 | | Crystal Structure of the C-terminal Periplasmic Domain of Phosphoethanolamine Transferase EptC from Campylobacter jejuni | | Descriptor: | UPF0141 protein yjdB, ZINC ION | | Authors: | Fage, C.D, Brown, D, Boll, J.M, Keatinge-Clay, A.T, Trent, M.S. | | Deposit date: | 2014-06-02 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7OYQ
 
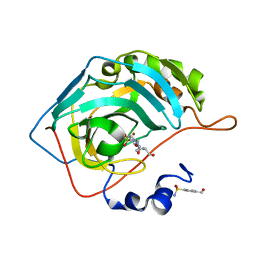 | | Carbonic anhydrase II in complex with Hit3-t2 (MH174) | | Descriptor: | Carbonic anhydrase 2, Hit3-t2 (MH174), ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2021-06-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
8EXL
 
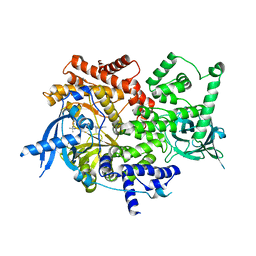 | | Crystal structure of PI3K-alpha in complex with taselisib | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXO
 
 | | Crystal structure of PI3K-alpha in complex with compound 19 | | Descriptor: | 1-{(4S,11aM)-2-[(4R)-2-oxo-4-(propan-2-yl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-prolinamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXU
 
 | | Crystal structure of PI3K-alpha in complex with compound 30 | | Descriptor: | (2S)-2-cyclopropyl-2-({(4S,11aM)-2-[(4S)-2-oxo-4-(trifluoromethyl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}amino)acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
4TQJ
 
 | | Structural basis of specific recognition of non-reducing terminal N-acetylglucosamine by an Agrocybe aegerita lection | | Descriptor: | Lectin 2 | | Authors: | Hu, Y.L, Ren, X.M, Li, D.F, Jiang, S, Lan, X.Q, Sun, H, Wang, D.C. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Specific Recognition of Non-Reducing Terminal N-Acetylglucosamine by an Agrocybe aegerita Lectin.
Plos One, 10, 2015
|
|
