2VVL
 
 | | The structure of MAO-N-D3, a variant of monoamine oxidase from Aspergillus niger. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N | | Authors: | Atkin, K.E, Hart, S, Turkenburg, J.P, Brzozowski, A.M, Grogan, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of Monoamine Oxidase from Aspergillus Niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution.
J.Mol.Biol., 384, 2008
|
|
7QFW
 
 | | S.c. Condensin peripheral Ycg1 subcomplex bound to DNA | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Synthetic DNA ligand, ... | | Authors: | Lecomte, L, Hassler, M, Haering, C, Eustermann, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | A hold-and-feed mechanism drives directional DNA loop extrusion by condensin.
Science, 376, 2022
|
|
8UQZ
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Gadolinium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GADOLINIUM ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2BFI
 
 | | Molecular basis for amyloid fibril formation and stability | | Descriptor: | SYNTHETIC PEPTIDE | | Authors: | Makin, O.S, Atkins, E, Sikorski, P, Johansson, J, Serpell, L.C. | | Deposit date: | 2004-12-07 | | Release date: | 2005-01-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Basis for Amyloid Fibril Formation and Stability
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7Q7T
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[(2S)-2,7-dimethyl-5,6-bis(oxidanylidene)-2,3-dihydro-1H-[1,4]oxazepino[6,5-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
4GOH
 
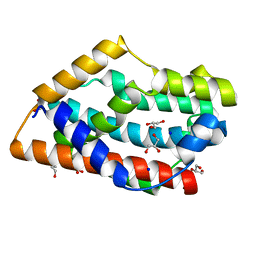 | |
1PAF
 
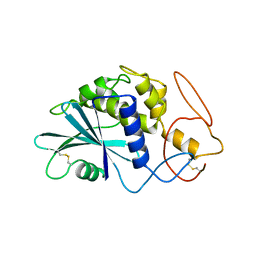 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
8V8I
 
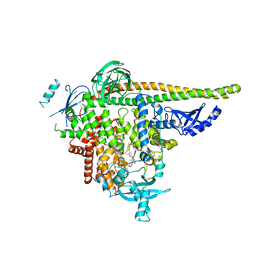 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket (compound 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, CHLORIDE ION, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
4N1H
 
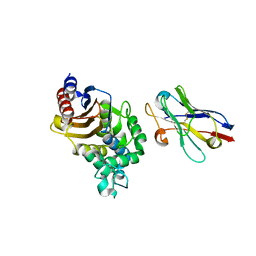 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4GOM
 
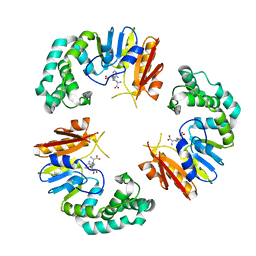 | |
7QK0
 
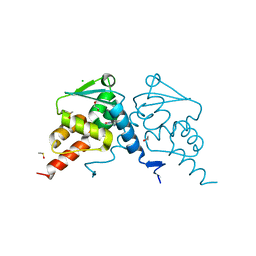 | | Crystal structure of human BCL6 BTB domain in complex with compound 12a | | Descriptor: | (2~{S})-10-[[5-chloranyl-2-[(3~{S},5~{R})-3-methyl-5-oxidanyl-piperidin-1-yl]pyrimidin-4-yl]amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Improved Binding Affinity and Pharmacokinetics Enable Sustained Degradation of BCL6 In Vivo .
J.Med.Chem., 65, 2022
|
|
2WDZ
 
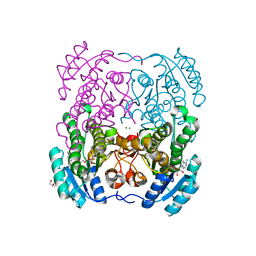 | | Crystal structure of the short chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD+ and 1,2-Pentandiol | | Descriptor: | (2S)-pentane-1,2-diol, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
5CIE
 
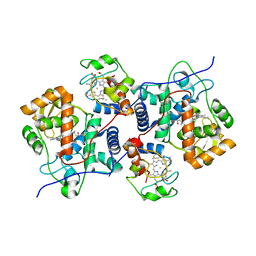 | |
4GPF
 
 | |
7QPX
 
 | |
8VA1
 
 | |
2BPU
 
 | | The Kedge Holmium Derivative of Hen Egg-White Lysozyme at high resolution from Single Wavelength Anomalous Diffraction | | Descriptor: | CHLORIDE ION, HOLMIUM ATOM, LYSOZYME C, ... | | Authors: | Jakoncic, J, Di Michiel, M, Zhong, Z, Honkimaki, V, Jouanneau, Y, Stojanoff, V. | | Deposit date: | 2005-04-25 | | Release date: | 2006-08-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Anomalous Diffraction at Ultra-High Energy for Protein Crystallography.
J.Appl.Crystallogr., 39, 2006
|
|
7QV6
 
 | | Amyloid fibril from the antimicrobial peptide aurein 3.3 | | Descriptor: | Aurein-3.3 | | Authors: | Buecker, R, Seuring, C, Cazey, C, Veith, K, Garcia-Alai, M, Gruenewald, K, Landau, M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The Cryo-EM structures of two amphibian antimicrobial cross-beta amyloid fibrils.
Nat Commun, 13, 2022
|
|
1PHG
 
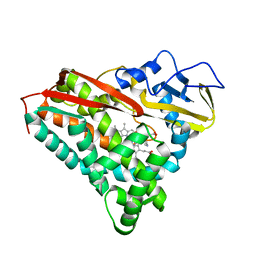 | |
7QV5
 
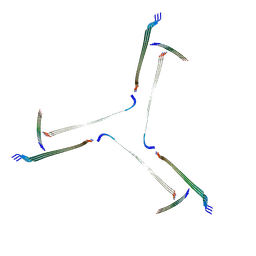 | | Amyloid fibril from the antimicrobial peptide uperin 3.5 | | Descriptor: | Uperin-3.5 | | Authors: | Buecker, R, Seuring, C, Cazey, C, Veith, K, Garcia-Alai, M, Gruenewald, K, Landau, M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-06-29 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Cryo-EM structures of two amphibian antimicrobial cross-beta amyloid fibrils.
Nat Commun, 13, 2022
|
|
7Q69
 
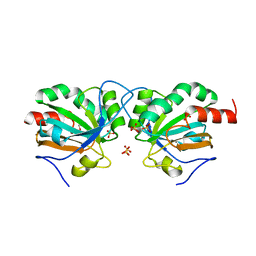 | | Crystal structure of Chaetomium thermophilum C30S Ahp1 in the pre-reaction state | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin domain-containing protein | | Authors: | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | Deposit date: | 2021-11-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
7KV0
 
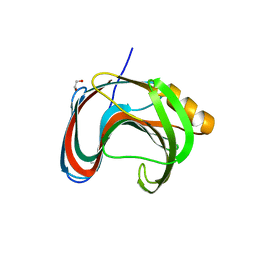 | | Crystallographic structure of Paenibacillus xylanivorans GH11 | | Descriptor: | 1,2-ETHANEDIOL, Endo-1,4-beta-xylanase | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and molecular dynamics investigations of ligand stabilization via secondary binding site interactions in Paenibacillus xylanivorans GH11 xylanase.
Comput Struct Biotechnol J, 19, 2021
|
|
7QWZ
 
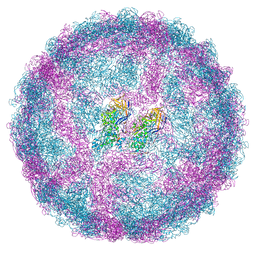 | | Full capsid of Saccharomyces cerevisiae virus L-BCLa | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
8U4C
 
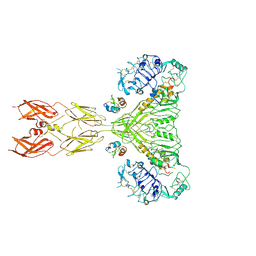 | | Cryo-EM structure of long form insulin receptor (IR-B) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin receptor, Insulin-like growth factor II | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
2YI8
 
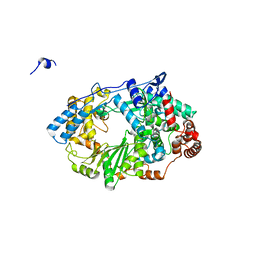 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | CHLORIDE ION, POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
