7TJO
 
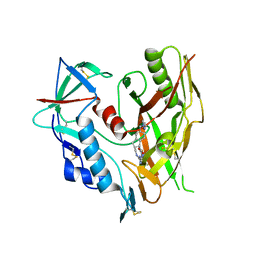 | | HIV-1 gp120 complex with CJF-II-197-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural and Functional Characterization of Indane-Core CD4-Mimetic Compounds Substituted with Heterocyclic Amines
ACS Medicinal Chemistry Letters, 14, 2023
|
|
6HSN
 
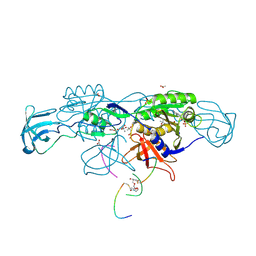 | | Crystal structure of the ternary complex of GephE-ADP-GABA(A) receptor derived peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,1'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis(1H-pyrrole-2,5-dione), ACETATE ION, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
5DGZ
 
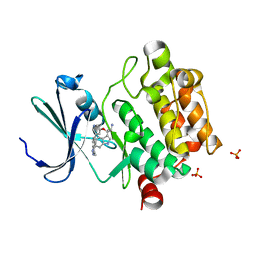 | | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2015-08-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1U27
 
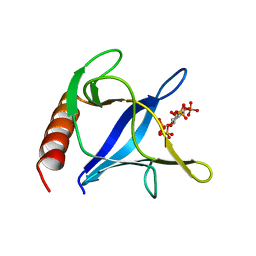 | | Triglycine variant of the ARNO Pleckstrin Homology Domain in complex with Ins(1,3,4,5)P4 | | Descriptor: | Cytohesin 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Cronin, T.C, DiNitto, J.P, Czech, M.P, Lambright, D.G. | | Deposit date: | 2004-07-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains
Embo J., 23, 2004
|
|
6JTU
 
 | | Crystal structure of MHETase from Ideonella sakaiensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Sagong, H.-Y, Seo, H, Kim, K.-J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decomposition of PET film by MHETase using Exo-PETase function
Acs Catalysis, 10, 2020
|
|
4Z2C
 
 | | Quinolone(Moxifloxacin)-DNA cleavage complex of gyrase from S. pneumoniae | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
4J1Z
 
 | |
5GUB
 
 | | Crystal structure of solute-binding protein complexed with sulfate group-free unsaturated chondroitin disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-08-26 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
5DIG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoromethyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7DT1
 
 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
6JVP
 
 | | Crystal structure of human MTH1 in complex with compound MI1024 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
5DI7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with an methyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5K6J
 
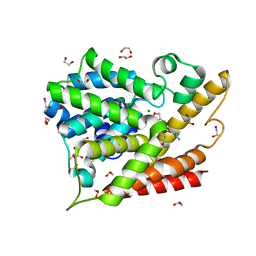 | | Human Phospodiesterase 4B in complex with pyridyloxy-benzoxaborole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-[[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl]oxy]-5-chloranyl-2-(4-oxidanylidenepentoxy)pyridine-3-carbonitrile, MAGNESIUM ION, ... | | Authors: | Rock, F.L, Zhou, Y, Sullivan, D. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | To be published
To Be Published
|
|
5TAX
 
 | | Structure of rabbit RyR1 (ryanodine dataset, class 1) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6EO7
 
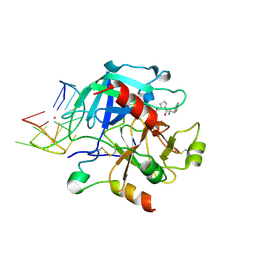 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA68B2 - MODIFIED HUMAN THROMBIN BINDING APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
3I2I
 
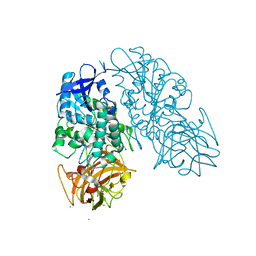 | | Cocaine Esterase with mutation T172R, bound to DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
4Z2I
 
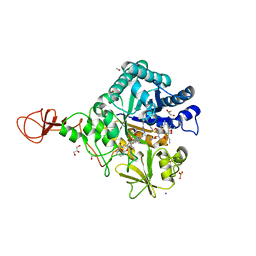 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 30 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
7WHD
 
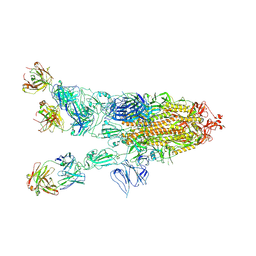 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Wang, X.W, Ge, J.W, Wang, Z.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
2WLZ
 
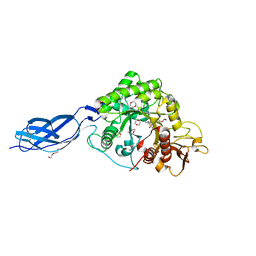 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
9GOI
 
 | | StmPr1, Stenotrophomonas maltophilia Protease 1, 36 kDa alkine serine protease in complex with Bortezomib | | Descriptor: | Alkaline serine protease, CALCIUM ION, GLYCEROL, ... | | Authors: | Sommer, M, Outzen, L, Negm, A, Windhorst, S, Weber, W, Betzel, C. | | Deposit date: | 2024-09-05 | | Release date: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Unveiling the structure, function and dynamics of StmPr1 in Stenotrophomonas maltophilia virulence.
Sci Rep, 15, 2025
|
|
4ZBK
 
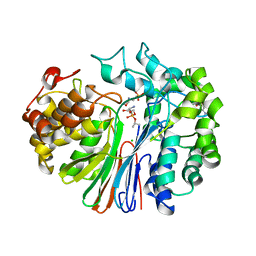 | | Crystal Structure of human GGT1 in complex with GGsTop inhibitor | | Descriptor: | (2S)-2-amino-4-[(S)-hydroxy(methoxy)phosphoryl]butanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Terzyan, S, Hanigan, M. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Human gamma-Glutamyl Transpeptidase 1: STRUCTURES OF THE FREE ENZYME, INHIBITOR-BOUND TETRAHEDRAL TRANSITION STATES, AND GLUTAMATE-BOUND ENZYME REVEAL NOVEL MOVEMENT WITHIN THE ACTIVE SITE DURING CATALYSIS.
J.Biol.Chem., 290, 2015
|
|
5DLD
 
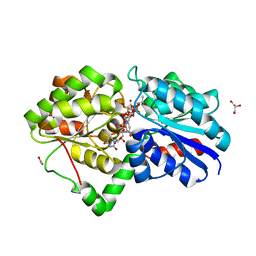 | |
1MO1
 
 | |
4ZAL
 
 | | Structure of UbiX E49Q mutant in complex with reduced FMN and dimethylallyl monophosphate | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dimethylallyl monophosphate, THIOCYANATE ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
6EEV
 
 | | Structure of class II HMG-CoA reductase from Delftia acidovorans with mevalonate bound | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl coenzyme A reductase, GLYCEROL, ... | | Authors: | Ragwan, E.R, Arai, E, Kung, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
