1GUI
 
 | | CBM4 structure and function | | Descriptor: | CALCIUM ION, GLYCEROL, LAMINARINASE 16A, ... | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-27 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1HG8
 
 | | Endopolygalacturonase from the phytopathogenic fungus Fusarium moniliforme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOPOLYGALACTURONASE | | Authors: | Federici, L, Caprari, C, Mattei, B, Savino, C, De Lorenzo, G, Cervone, F, Tsernoglou, D. | | Deposit date: | 2000-12-13 | | Release date: | 2001-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Requirements of Endopolygalacturonase for the Interaction with Pgip (Polygalacturonase-Inhibiting Protein)
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HGD
 
 | | BINDING OF INFLUENZA VIRUS HEMAGGLUTININ TO ANALOGS OF ITS CELL-SURFACE RECEPTOR, SIALIC ACID: ANALYSIS BY PROTON NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Sauter, N.K, Hanson, J.E, Glick, G.D, Brown, J.H, Crowther, R.L, Park, S.-J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1991-11-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: analysis by proton nuclear magnetic resonance spectroscopy and X-ray crystallography.
Biochemistry, 31, 1992
|
|
4DI9
 
 | | CRYSTAL STRUCTURE OF THE D248A mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 6.5 | | Descriptor: | (1E,3Z)-4-hydroxybuta-1,3-diene-1,2,4-tricarboxylic acid, 2-pyrone-4,6-dicarbaxylate hydrolase, ACETATE ION | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
1H3J
 
 | | STRUCTURE OF RECOMBINANT COPRINUS CINEREUS PEROXIDASE DETERMINED TO 2.0 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Petersen, J.F.W, Houborg, K, Harris, P, Larsen, S. | | Deposit date: | 2002-09-05 | | Release date: | 2003-06-12 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impact of the Physical and Chemical Environment on the Molecular Structure of Coprinus Cinereus Peroxidase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1GSL
 
 | |
4DM0
 
 | | TN5 transposase: 20MER OUTSIDE END 2 MN complex | | Descriptor: | 1,2-ETHANEDIOL, DNA NON-TRANSFERRED STRAND, DNA TRANSFERRED STRAND, ... | | Authors: | Klenchin, V.A, Lovell, S, Goryshin, I.Y, Reznikoff, W.R, Rayment, I. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-metal active site binding of a TN5 transposase synaptic complex
Nat.Struct.Biol., 9, 2002
|
|
1D7C
 
 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, PH 4.6 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1D7B
 
 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, PH 7.5 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1CEN
 
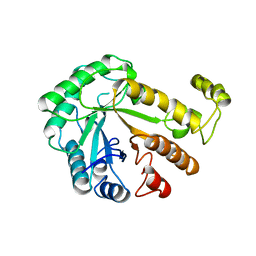 | |
6LCO
 
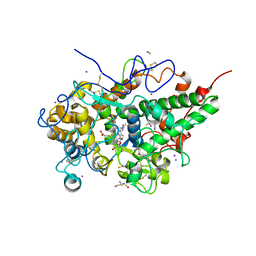 | | Crystal structure of bovine lactoperoxidase with substrates thiocynate and iodide bound at the distal heme side at 1.99 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Sirohi, H.V, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of bovine lactoperoxidase with substrates thiocynate and iodide bound at the distal heme side at 1.99 A resolution.
To Be Published
|
|
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
6APB
 
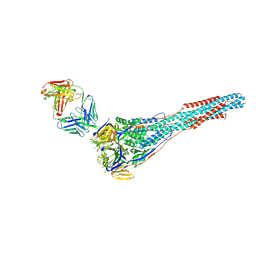 | |
1D1I
 
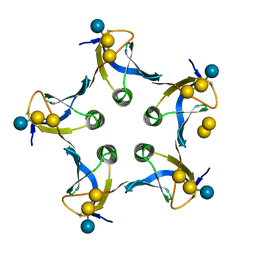 | |
4KJG
 
 | | Structure of Rat Intestinal Alkaline Phosphatase expressed in insect cell | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ghosh, K, Anumula, R.K, Laksmaiah, B.K. | | Deposit date: | 2013-05-03 | | Release date: | 2013-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of rat intestinal alkaline phosphatase - Role of crown domain in mammalian alkaline phosphatases.
J.Struct.Biol., 184, 2013
|
|
4KMZ
 
 | | Human folate receptor beta (FOLR2) in complex with the folate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FOLIC ACID, ... | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1CF3
 
 | | GLUCOSE OXIDASE FROM APERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (GLUCOSE OXIDASE), ... | | Authors: | Hecht, H.J, Kalisz, H. | | Deposit date: | 1999-03-23 | | Release date: | 1999-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CGY
 
 | |
4DIA
 
 | | CRYSTAL STRUCTURE OF THE D248N mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 4.6 | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
3S9M
 
 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, cryocooled 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6AZP
 
 | |
1GZ7
 
 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
4M7F
 
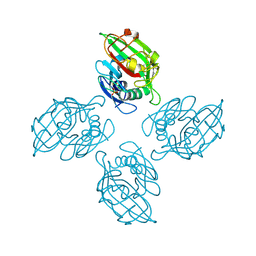 | | Crystal structure of tetrameric fibrinogen-like recognition domain of FIBCD1 with bound ManNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Shrive, A.K, Greenhough, T.J, Holmskov, U. | | Deposit date: | 2013-08-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Tetrameric Fibrinogen-like Recognition Domain of Fibrinogen C Domain Containing 1 (FIBCD1) Protein.
J.Biol.Chem., 289, 2014
|
|
1HHC
 
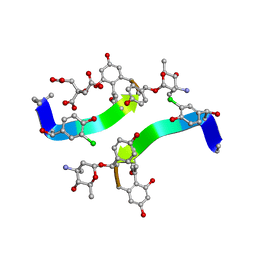 | | Crystal structure of Decaplanin - space group P21, second form | | Descriptor: | 4-epi-vancosamine, CITRIC ACID, DECAPLANIN, ... | | Authors: | Lehmann, C, Vertessy, L, Sheldrick, G.M, Dauter, Z, Dauter, M. | | Deposit date: | 2000-12-22 | | Release date: | 2005-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structures of Four Crystal Forms of Decaplanin
Helv.Chim.Acta, 86, 2003
|
|
