1CMV
 
 | | HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | HUMAN CYTOMEGALOVIRUS PROTEASE | | Authors: | Shieh, H.-S, Kurumbail, R.G, Stevens, A.M, Stegeman, R.A, Sturman, E.J, Pak, J.Y, Wittwer, A.J, Palmier, M.O, Wiegand, R.C, Holwerda, B.C, Stallings, W.C. | | Deposit date: | 1996-08-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Three-dimensional structure of human cytomegalovirus protease.
Nature, 383, 1996
|
|
1CQY
 
 | | STARCH BINDING DOMAIN OF BACILLUS CEREUS BETA-AMYLASE | | Descriptor: | BETA-AMYLASE | | Authors: | Yoon, H.J, Hirata, A, Adachi, M, Sekine, A, Utsumi, S, Mikami, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Separated Starch-Binding Domain of Bacillus cereus B-amylase
To be Published
|
|
1CG6
 
 | |
1CGQ
 
 | |
1CVS
 
 | | CRYSTAL STRUCTURE OF A DIMERIC FGF2-FGFR1 COMPLEX | | Descriptor: | FIBROBLAST GROWTH FACTOR 2, FIBROBLAST GROWTH FACTOR RECEPTOR 1, SULFATE ION | | Authors: | Plotnikov, A.N, Schlessinger, J, Hubbard, S.R, Mohammadi, M. | | Deposit date: | 1999-08-24 | | Release date: | 2000-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for FGF receptor dimerization and activation.
Cell(Cambridge,Mass.), 98, 1999
|
|
1D1P
 
 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
1CYN
 
 | |
1D5E
 
 | |
1CZE
 
 | | ASPARTATE AMINOTRANSFERASE MUTANT ATB17/139S/142N WITH SUCCINIC ACID | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SUCCINIC ACID | | Authors: | Okamoto, A, Oue, S, Yano, T, Kagamiyama, H. | | Deposit date: | 1999-09-02 | | Release date: | 2000-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cocrystallization of a mutant aspartate aminotransferase with a C5-dicarboxylic substrate analog: structural comparison with the enzyme-C4-dicarboxylic analog complex.
J.Biochem.(Tokyo), 127, 2000
|
|
1CZL
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1CZV
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: DIMERIC CRYSTAL FORM | | Descriptor: | PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
1D7M
 
 | | COILED-COIL DIMERIZATION DOMAIN FROM CORTEXILLIN I | | Descriptor: | CORTEXILLIN I | | Authors: | Burkhard, P, Kammerer, R.A, Steinmetz, M.O, Bourenkov, G.P, Aebi, U. | | Deposit date: | 1999-10-19 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges.
Structure Fold.Des., 8, 2000
|
|
1CWB
 
 | |
1D8Y
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA | | Descriptor: | D(T)19 OLIGOMER, DNA POLYMERASE I, SULFATE ION, ... | | Authors: | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | Deposit date: | 1999-10-26 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CZJ
 
 | | CYTOCHROME C OF CLASS III (AMBLER) 26 KD | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Czjzek, M, Haser, R. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a dimeric octaheme cytochrome c3 (M(r) 26,000) from Desulfovibrio desulfuricans Norway.
Structure, 4, 1996
|
|
1CE0
 
 | |
1D3R
 
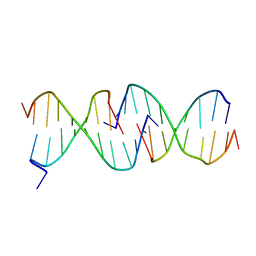 | | CRYSTAL STRUCTURE OF TRIPLEX DNA | | Descriptor: | DNA (5'-D(*CP*(BRU)P*CP*CP*(BRU)P*CP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*CP*GP*GP*AP*G)-3') | | Authors: | Rhee, S, Han, Z.-J, Liu, K, Todd Miles, H.T, Davies, D.R. | | Deposit date: | 1999-09-30 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a triple helical DNA with a triplex-duplex junction.
Biochemistry, 38, 1999
|
|
1CE9
 
 | | HELIX CAPPING IN THE GCN4 LEUCINE ZIPPER | | Descriptor: | PROTEIN (GCN4-PMSE) | | Authors: | Lu, M, Shu, W, Ji, H, Spek, E, Wang, L.-Y, Kallenbach, N.R. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Helix capping in the GCN4 leucine zipper.
J.Mol.Biol., 288, 1999
|
|
1CGS
 
 | |
1D5Z
 
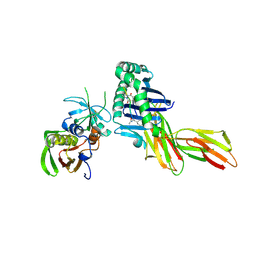 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | PROTEIN (ENTEROTOXIN TYPE B), PROTEIN (HLA CLASS II HISTOCOMPATIBILITY ANTIGEN), PROTEIN (PEPTIDOMIMETIC INHIBITOR) | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
1D6P
 
 | | HUMAN LYSOZYME L63 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL N,N'-DIACETYLCHITOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LYSOZYME | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
2QRW
 
 | | Crystal structure of Mycobacterium tuberculosis trHbO WG8F mutant | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbO, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Milani, M, Bolognesi, M. | | Deposit date: | 2007-07-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Roles of Tyr(CD1) and Trp(G8) in Mycobacterium tuberculosis Truncated Hemoglobin O in Ligand Binding and on the Heme Distal Site Architecture
Biochemistry, 46, 2007
|
|
1CHN
 
 | |
3T6W
 
 | | Crystal Structure of Steccherinum ochraceum Laccase obtained by multi-crystals composite data collection technique (10% dose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I, Kolomytseva, M, Golovleva, L, Scozzafava, A, Chernykh, A.M. | | Deposit date: | 2011-07-29 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reaction intermediates and redox state changes in a blue laccase from Steccherinum ochraceum observed by crystallographic high/low X-ray dose experiments.
J.Inorg.Biochem., 111, 2012
|
|
1CI0
 
 | | PNP OXIDASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (PNP OXIDASE) | | Authors: | Shi, W, Ostrov, D.A, Gerchman, S.E, Graziano, V, Kycia, H, Studier, B, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-04-06 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of PNP Oxidase from S. Cerevisiae
To be Published
|
|
