1GPO
 
 | |
1B52
 
 | |
28SP
 
 | |
1H0T
 
 | | An affibody in complex with a target protein: structure and coupled folding | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN A, ZSPA-1 AFFIBODY | | Authors: | Wahlberg, E, Lendel, C, Helgstrand, M, Allard, P, Dincbas-Renqvist, V, Hedqvist, A, Berglund, H, Nygren, P.-A, Hard, T. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Affibody in Complex with a Target Protein: Structure and Coupled Folding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1BDT
 
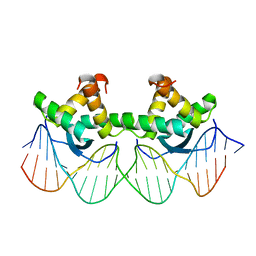 | | WILD TYPE GENE-REGULATING PROTEIN ARC/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (GENE-REGULATING PROTEIN ARC) | | Authors: | Schilbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | Deposit date: | 1998-05-11 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1H55
 
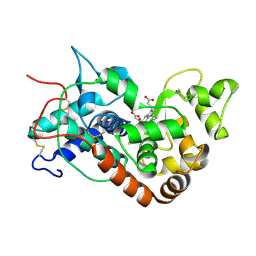 | | STRUCTURE OF HORSERADISH PEROXIDASE C1A COMPOUND II | | Descriptor: | ACETATE ION, CALCIUM ION, OXYGEN ATOM, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-18 | | Release date: | 2002-06-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1VBU
 
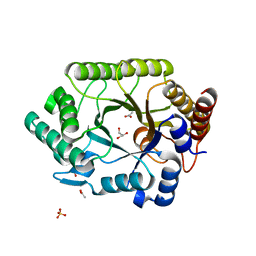 | | Crystal structure of native xylanase 10B from Thermotoga maritima | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | Deposit date: | 2004-03-02 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1BAM
 
 | |
248D
 
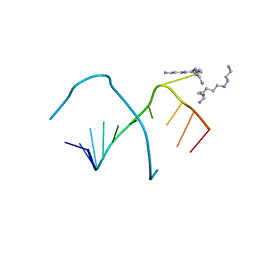 | | CRYSTAL STRUCTURES OF AN A-FORM DUPLEX WITH SINGLE-ADENOSINE BULGES AND A CONFORMATIONAL BASIS FOR SITE SPECIFIC RNA SELF-CLEAVAGE | | Descriptor: | DNA/RNA (5'-R(*GP*CP*GP*)-D(*AP*TP*AP*TP*AP*)-R(*CP*GP*C)-3'), ORTHORHOMBIC, SPERMINE | | Authors: | Portmann, S, Grimm, S, Workman, C, Usman, N, Egli, M. | | Deposit date: | 1996-02-02 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of an A-form duplex with single-adenosine bulges and a conformational basis for site-specific RNA self-cleavage.
Chem.Biol., 3, 1996
|
|
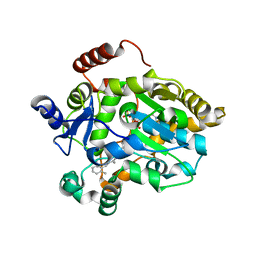
1B32
 
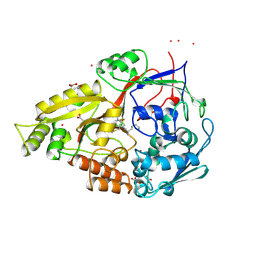 | |
1FML
 
 | | CRYSTAL STRUCTURE OF RETINOL DEHYDRATASE IN A COMPLEX WITH RETINOL AND PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, RETINOL, ... | | Authors: | Pakhomova, S, Kobayashi, M, Buck, J, Newcomer, M.E. | | Deposit date: | 2000-08-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A helical lid converts a sulfotransferase to a dehydratase.
Nat.Struct.Biol., 8, 2001
|
|
1B3S
 
 | |
1B2S
 
 | |
1B3F
 
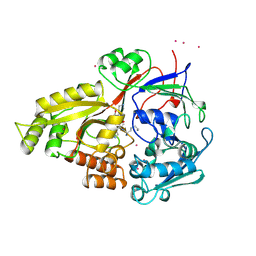 | |
1B8I
 
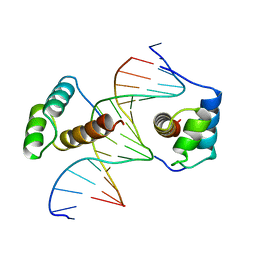 | | STRUCTURE OF THE HOMEOTIC UBX/EXD/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*GP*AP*TP*TP*TP*AP*TP*GP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*CP*AP*TP*AP*AP*AP*TP*CP*AP*C)-3'), PROTEIN (HOMEOBOX PROTEIN EXTRADENTICLE), ... | | Authors: | Passner, J.M, Ryoo, H.-D, Shen, L, Mann, R.S, Aggarwal, A.K. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex.
Nature, 397, 1999
|
|
1B5I
 
 | |
1FRP
 
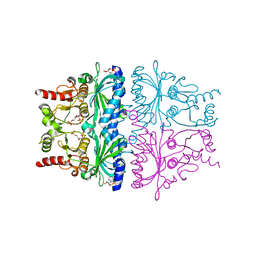 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-08-26 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1FDG
 
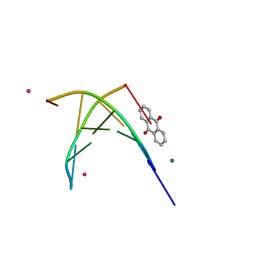 | | BINDING OF A MACROCYCLIC BISACRIDINE AND AMETANTRONE TO CGTACG INVOLVES SIMILAR UNUSUAL INTERCALATION PLATFORMS (AMETANTRONE COMPLEX) | | Descriptor: | 1,4-BIS-[2-(2-HYDROXY-ETHYLAMINO)-ETHYLAMINO]-ANTHRAQUINONE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Yang, X.-L, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of a macrocyclic bisacridine and ametantrone to CGTACG involves similar unusual intercalation platforms.
Biochemistry, 39, 2000
|
|
1FE6
 
 | |
1FJ4
 
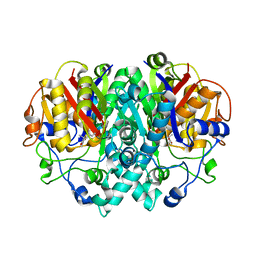 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH THIOLACTOMYCIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I, THIOLACTOMYCIN | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
1CJX
 
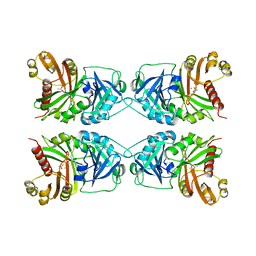 | | CRYSTAL STRUCTURE OF PSEUDOMONAS FLUORESCENS HPPD | | Descriptor: | 4-HYDROXYPHENYLPYRUVATE DIOXYGENASE, ACETATE ION, ETHYL MERCURY ION, ... | | Authors: | Serre, L, Sailland, A, Sy, D, Boudec, P, Rolland, A, Pebay-Peroulla, E, Cohen-Addad, C. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway.
Structure Fold.Des., 7, 1999
|
|
1KTH
 
 | |
1XII
 
 | |
1XIJ
 
 | |
1XEI
 
 | |
