9FOJ
 
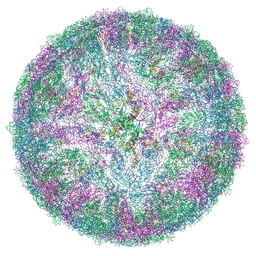 | | LGTV TP21. Langat virus, strain TP21 | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-06-12 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | The influence of the pre-membrane and envelope proteins on structure, pathogenicity and tropism of tick-borne encephalitis virus
Biorxiv, 2024
|
|
4ZP2
 
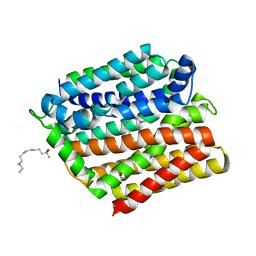 | | Crystal structure of E. coli multidrug transporter MdfA in complex with n-dodecyl-N,N-dimethylamine-N-oxide | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Multidrug transporter MdfA | | Authors: | Zhang, X.C, Heng, J, Zhao, Y, Wang, X. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-bound structure of the E. coli multidrug resistance transporter MdfA
Cell Res., 25, 2015
|
|
3C9C
 
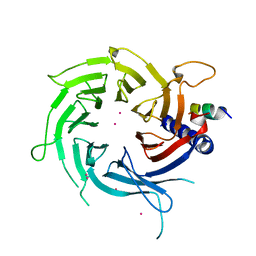 | | Structural Basis of Histone H4 Recognition by p55 | | Descriptor: | CADMIUM ION, Chromatin assembly factor 1 p55 subunit, Histone H4, ... | | Authors: | Song, J.J, Garlick, J.D, Kingston, R.E. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of histone H4 recognition by p55.
Genes Dev., 22, 2008
|
|
9BRB
 
 | |
9BRD
 
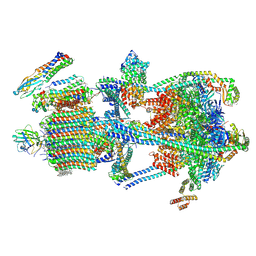 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3 | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9BRC
 
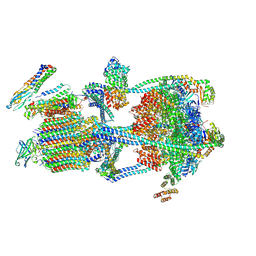 | |
4W6W
 
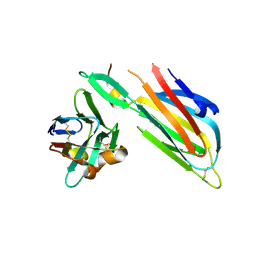 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF6 | | Descriptor: | F18 fimbrial adhesin AC, NbFedF6 | | Authors: | Moonens, K, De Kerpel, M, Annelies, C, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
3CFS
 
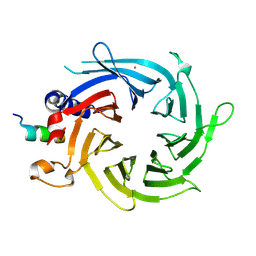 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4, Histone-binding protein RBBP7 | | Authors: | Murzina, N.V, Pei, X.-Y, Pratap, J.V, Sparkes, M, Vicente-Garcia, J, Ben-Shahar, T.R, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
4W6X
 
 | | Co-complex structure of the lectin domain of F18 fimbrial adhesin FedF with inhibitory nanobody NbFedF7 | | Descriptor: | F18 fimbrial adhesin AC, Nanobody NbFedF7 | | Authors: | Moonens, K, De Kerpel, M, Coddens, A, Cox, E, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Nanobody Mediated Inhibition of Attachment of F18 Fimbriae Expressing Escherichia coli.
Plos One, 9, 2014
|
|
4X9E
 
 | | DEOXYGUANOSINETRIPHOSPHATE TRIPHOSPHOHYDROLASE from Escherichia coli with two DNA effector molecules | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, RNA (5'-R(P*CP*CP*C)-3') | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
3CG5
 
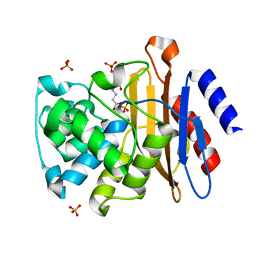 | |
7NPR
 
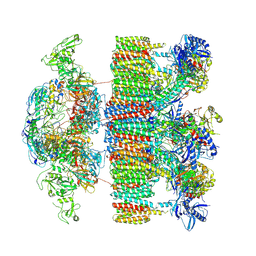 | | Structure of an intact ESX-5 inner membrane complex, Composite C3 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
9BQ4
 
 | |
7NP7
 
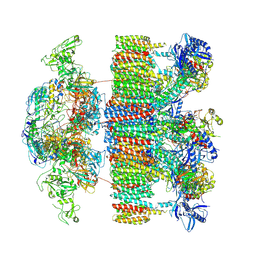 | | Structure of an intact ESX-5 inner membrane complex, Composite C1 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
5YFI
 
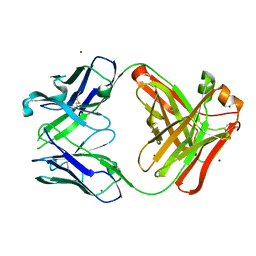 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
3JZ7
 
 | |
4UHS
 
 | |
4WA9
 
 | |
4UM4
 
 | |
5YHL
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | Descriptor: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
4UHT
 
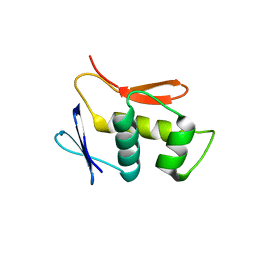 | |
5A24
 
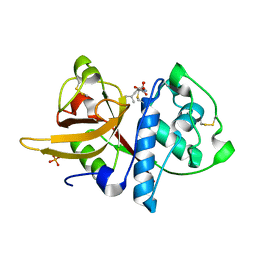 | | Crystal structure of Dionain-1, the major endopeptidase in the Venus flytrap digestive juice | | Descriptor: | DIONAIN-1, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, PHOSPHATE ION | | Authors: | Risor, M.W, Thomsen, L.R, Sanggaard, K.W, Nielsen, T.A, Thogersen, I.B, Lukassen, M.V, Rossen, L, Garcia-Ferrer, I, Guevara, T, Meinjohanns, E, Nielsen, N.C, Gomis-Ruth, F.X, Enghild, J.J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enzymatic and Structural Characterization of the Major Endopeptidase in the Venus Flytrap Digestion Fluid.
J.Biol.Chem., 291, 2016
|
|
9FMT
 
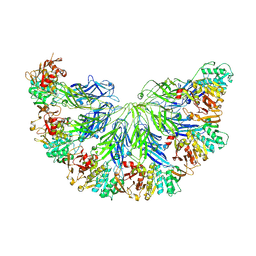 | |
9FP2
 
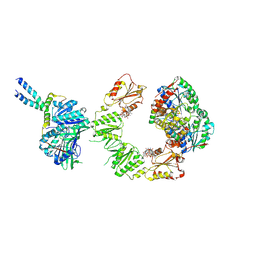 | | Cryo-EM structure of the BcsEFRQ regulatory subcomplex for E. coli cellulose secretion in non-saturating c-di-GMP (local) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Cell division protein, ... | | Authors: | Anso, I, Krasteva, P.V. | | Deposit date: | 2024-06-12 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Nat Commun, 15, 2024
|
|
9FP0
 
 | | Cryo-EM structure of the 'crown'less Bcs macrocomplex for E. coli cellulose secretion in non-saturating c-di-GMP (local) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Cell division protein, ... | | Authors: | Anso, I, Krasteva, P.V. | | Deposit date: | 2024-06-12 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Nat Commun, 15, 2024
|
|
