4OYE
 
 | | Crystal structure of GltPh R397A in apo | | Descriptor: | 425aa long hypothetical proton glutamate symport protein | | Authors: | Boudker, O, Oh, S. | | Deposit date: | 2014-02-11 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Elife, 3, 2014
|
|
4OIR
 
 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4O9L
 
 | |
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OLT
 
 | | Chitosanase complex structure | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase, GLYCEROL | | Authors: | Liu, W.Z, Lyu, Q.Q, Han, B.Q. | | Deposit date: | 2014-01-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the substrate-binding mechanism for a novel chitosanase.
Biochem.J., 461, 2014
|
|
4MEX
 
 | | Crystal structure of Escherichia coli RNA polymerase in complex with salinamide A | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
4M8M
 
 | |
4OIN
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4O9F
 
 | |
4OIO
 
 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
6A9V
 
 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9U
 
 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9T
 
 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 58 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
5YZV
 
 | | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA | | Descriptor: | Probable serine/threonine-protein kinase PkwA | | Authors: | Li, D.Y, Shen, C, Du, Y, Qiao, F.F, Kong, T, Yuan, L.R, Zhang, D.L, Wu, X.H, Wu, Y.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biophysical and structural characterization of the thermostable WD40 domain of a prokaryotic protein, Thermomonospora curvata PkwA
Sci Rep, 8, 2018
|
|
5CFY
 
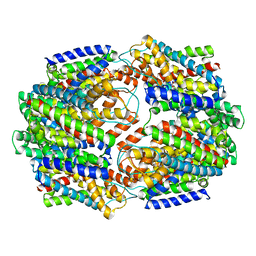 | | CRYSTAL STRUCTURE OF GLTPH R397A IN COMPLEX WITH NA+ AND L-ASP | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, SODIUM ION | | Authors: | Boudker, O, Oh, S. | | Deposit date: | 2015-07-08 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Elife, 3, 2014
|
|
5ZT0
 
 | |
5ZQV
 
 | |
5Z5O
 
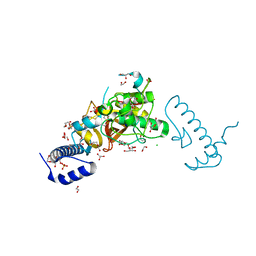 | | Structure of Pycnonodysostosis disease related I249T mutant of human cathepsin K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Biswas, S, Roy, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Not all pycnodysostosis-related mutants of human cathepsin K are inactive - crystal structure and biochemical studies of an active mutant I249T.
FEBS J., 285, 2018
|
|
4PPS
 
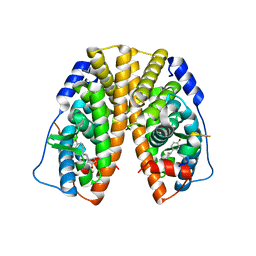 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with an A-CD ring estrogen derivative | | Descriptor: | (1S,3aR,5R,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
4P1A
 
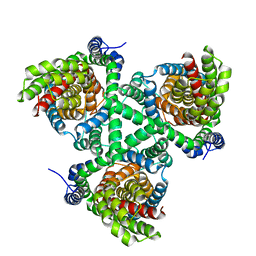 | |
4P19
 
 | |
4P3J
 
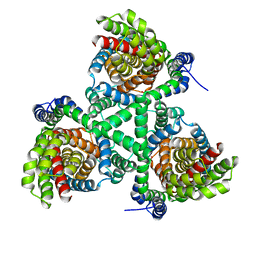 | |
4P0T
 
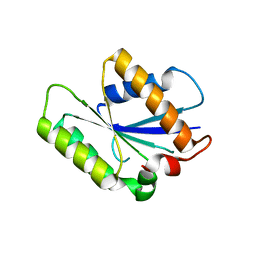 | |
4PP6
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Resveratrol | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, RESVERATROL | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-26 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
