8HCR
 
 | |
4P4Q
 
 | |
6P41
 
 | |
8GY3
 
 | | Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Cytochrome c subunit of aldehyde dehydrogenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
8GY2
 
 | | Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans | | Descriptor: | Alcohol dehydrogenase (quinone), cytochrome c subunit, dehydrogenase subunit, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
6P43
 
 | |
6P42
 
 | |
4XYD
 
 | | Nitric oxide reductase from Roseobacter denitrificans (RdNOR) | | Descriptor: | CALCIUM ION, COPPER (II) ION, FE (III) ION, ... | | Authors: | Crow, A. | | Deposit date: | 2015-02-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Membrane-intrinsic Nitric Oxide Reductase from Roseobacter denitrificans.
Biochemistry, 55, 2016
|
|
8JEK
 
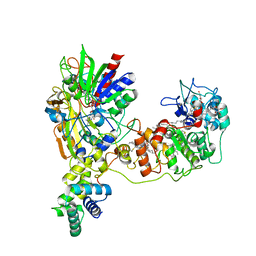 | | Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
8JEJ
 
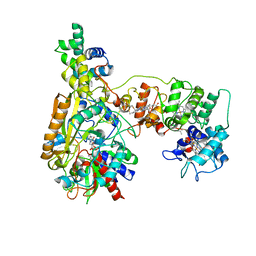 | | Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
5JUY
 
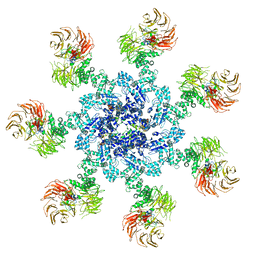 | | Active human apoptosome with procaspase-9 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Caspase-9, ... | | Authors: | Cheng, T.C, Hong, C, Akey, I.V, Yuan, S, Akey, C.W. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-19 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A near atomic structure of the active human apoptosome.
Elife, 5, 2016
|
|
6HWH
 
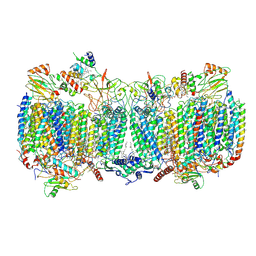 | | Structure of a functional obligate respiratory supercomplex from Mycobacterium smegmatis | | Descriptor: | CARDIOLIPIN, COPPER (II) ION, Co-purified unknown peptide built as polyALA, ... | | Authors: | Wiseman, B, Nitharwal, R.G, Fedotovskaya, O, Schafer, J, Guo, H, Kuang, Q, Benlekbir, S, Sjostrand, D, Adelroth, P, Rubinstein, J.L, Brzezinski, P, Hogbom, M. | | Deposit date: | 2018-10-12 | | Release date: | 2018-11-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a functional obligate complex III2IV2respiratory supercomplex from Mycobacterium smegmatis.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7W2J
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7VZR
 
 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
7VZG
 
 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
6H5L
 
 | |
7WSQ
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
5IY5
 
 | | Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, S, Baba, J, Aoe, S, Shimada, A, Yamashita, E, Tsukihara, T. | | Deposit date: | 2016-03-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
EMBO J., 36, 2017
|
|
8K6J
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-H1147A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (H1147A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8K6K
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (N1146A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
7ZS8
 
 | | Mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae at 1.4 Angstrom resolution | | Descriptor: | CALCIUM ION, Cytochrome-c peroxidase, HEME C, ... | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Characterization of Neisseria gonorrhoeae Bacterial Peroxidase-Insights into the Catalytic Cycle of Bacterial Peroxidases.
Int J Mol Sci, 24, 2023
|
|
4NFG
 
 | | K13R mutant of horse cytochrome c and yeast cytochrome c peroxidase complex | | Descriptor: | Cytochrome c, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Meulenbroek, E.M, Bashir, Q, Ubbink, M, Pannu, N.S. | | Deposit date: | 2013-10-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Engineering specificity in a dynamic protein complex with a single conserved mutation.
Febs J., 281, 2014
|
|
1S6V
 
 | | Structure of a cytochrome c peroxidase-cytochrome c site specific cross-link | | Descriptor: | Cytochrome c peroxidase, mitochondrial, Cytochrome c, ... | | Authors: | Guo, M, Bhaskar, B, Li, H, Barrows, T.P, Poulos, T.L. | | Deposit date: | 2004-01-27 | | Release date: | 2004-04-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and characterization of a cytochrome c peroxidase-cytochrome c site-specific cross-link
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2B0Z
 
 | |
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
