8DM7
 
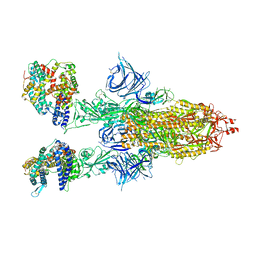 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DMA
 
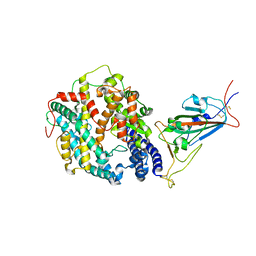 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM5
 
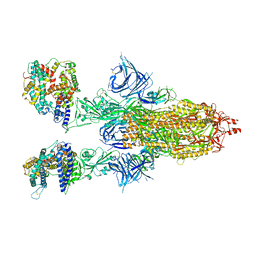 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM8
 
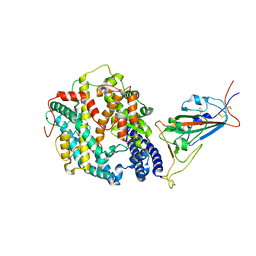 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM9
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
2OCC
 
 | |
2OT4
 
 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
1FS8
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-SULFATE COMPLEX | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FS7
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FDI
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI COMPLEXED WITH THE INHIBITOR NITRITE | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
1FFT
 
 | | The structure of ubiquinol oxidase from Escherichia coli | | Descriptor: | COPPER (II) ION, HEME O, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abramson, J, Riistama, S, Larsson, G, Jasaitis, A, Svensson-Ek, M, Puustinen, A, Iwata, S, Wikstrom, M. | | Deposit date: | 2000-07-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site.
Nat.Struct.Biol., 7, 2000
|
|
1HPI
 
 | | MOLECULAR STRUCTURE OF THE OXIDIZED HIGH-POTENTIAL IRON-SULFUR PROTEIN ISOLATED FROM ECTOTHIORHODOSPIRA VACUOLATA | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Benning, M.M, Meyer, T.E, Rayment, I, Holden, H.M. | | Deposit date: | 1993-12-09 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structure of the oxidized high-potential iron-sulfur protein isolated from Ectothiorhodospira vacuolata.
Biochemistry, 33, 1994
|
|
1HIP
 
 | | TWO-ANGSTROM CRYSTAL STRUCTURE OF OXIDIZED CHROMATIUM HIGH POTENTIAL IRON PROTEIN | | Descriptor: | HIGH POTENTIAL IRON PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Carterjunior, C.W, Kraut, J, Freer, S.T, Xuong, N.-H, Alden, R.A, Bartsch, R.G. | | Deposit date: | 1975-04-01 | | Release date: | 1976-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein.
J.Biol.Chem., 249, 1974
|
|
1HLQ
 
 | | CRYSTAL STRUCTURE OF RHODOFERAX FERMENTANS HIGH POTENTIAL IRON-SULFUR PROTEIN REFINED TO 1.45 A | | Descriptor: | HIGH-POTENTIAL IRON-SULFUR PROTEIN, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Gonzalez, A, Ciurli, S, Benini, S. | | Deposit date: | 2000-12-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Rhodoferax fermentans high-potential iron-sulfur protein solved by MAD.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1GU6
 
 | | Structure of the Periplasmic Cytochrome c Nitrite Reductase from Escherichia coli | | Descriptor: | CALCIUM ION, CYTOCHROME C552, GLYCEROL, ... | | Authors: | Bamford, V.A, Angove, H.C, Seward, H.E, Thomson, A.J, Cole, J.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Spectroscopy of the Periplasmic Cytochrome C Nitrite Reductase from Escherichia Coli
Biochemistry, 41, 2002
|
|
1FDO
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
4BC5
 
 | | Crystal structure of human D-xylulokinase in complex with inhibitor 5- deoxy-5-fluoro-D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, 5-deoxy-5-fluoro-D-xylulose, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
5NA4
 
 | | NADH:quinone oxidoreductase (NDH-II) from Staphylococcus aureus - E172S mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH dehydrogenase-like protein SAOUHSC_00878 | | Authors: | Brito, J.A, Athayde, D, Sousa, F.M, Sena, F.V, Pereira, M.M, Archer, M. | | Deposit date: | 2017-02-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The key role of glutamate 172 in the mechanism of type II NADH:quinone oxidoreductase of Staphylococcus aureus.
Biochim. Biophys. Acta, 1858, 2017
|
|
2RDZ
 
 | | High Resolution Crystal Structure of the Escherichia coli Cytochrome c Nitrite Reductase. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, RIchardson, D.J. | | Deposit date: | 2007-09-25 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
2RF7
 
 | | Crystal structure of the escherichia coli nrfa mutant Q263E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
4CY2
 
 | | Crystal structure of the KANSL1-WDR5-KANSL2 complex. | | Descriptor: | KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, KAT8 REGULATORY NSL COMPLEX SUBUNIT 2, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4BC4
 
 | | Crystal structure of human D-xylulokinase in complex with D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, D-XYLULOSE, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
2WDQ
 
 | | E. coli succinate:quinone oxidoreductase (SQR) with carboxin bound | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-25 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site.
J. Biol. Chem., 284, 2009
|
|
2WDV
 
 | | E. coli succinate:quinone oxidoreductase (SQR) with an empty quinone- binding pocket | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-26 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia Coli Succinate:Quinone Oxidoreductase with an Occupied and Empty Quinone- Binding Site.
J.Biol.Chem., 284, 2009
|
|
2WDR
 
 | | E. coli succinate:quinone oxidoreductase (SQR) with pentachlorophenol bound | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-25 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia Coli Succinate:Quinone Oxidoreductase with an Occupied and Empty Quinone- Binding Site.
J.Biol.Chem., 284, 2009
|
|
