7RY6
 
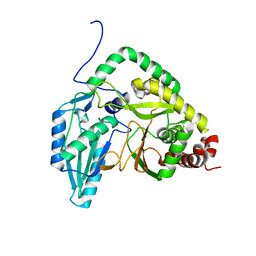 | | Solution NMR structural bundle of the first cyclization domain from yersiniabactin synthetase (Cy1) impacted by dynamics | | Descriptor: | HMWP2 nonribosomal peptide synthetase | | Authors: | Kancherla, A.K, Mishra, S.H, Marincin, K.A, Nerli, S, Sgourakis, N.G, Dowling, D.P, Bouvignies, G, Frueh, D.P. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global protein dynamics as communication sensors in peptide synthetase domains.
Sci Adv, 8, 2022
|
|
7R9X
 
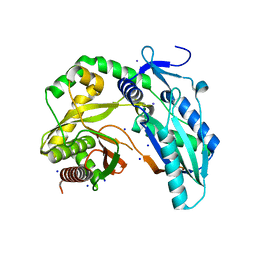 | |
1L5A
 
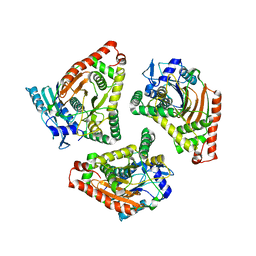 | | Crystal Structure of VibH, an NRPS Condensation Enzyme | | Descriptor: | amide synthase | | Authors: | Keating, T.A, Marshall, C.G, Walsh, C.T, Keating, A.E. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains.
Nat.Struct.Biol., 9, 2002
|
|
6TA8
 
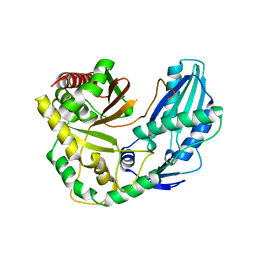 | |
6AEF
 
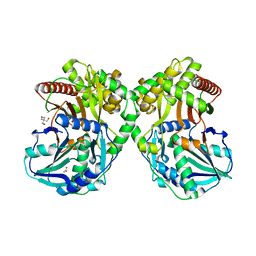 | | PapA2 acyl transferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Trehalose-2-sulfate acyltransferase PapA2, ... | | Authors: | Chaudhary, S, Rao, V, Panchal, V. | | Deposit date: | 2018-08-04 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A novel mutation alters the stability of PapA2 resulting in the complete abrogation of sulfolipids in clinical mycobacterial strains.
Faseb Bioadv, 1, 2019
|
|
6AD3
 
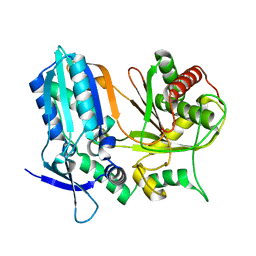 | |
4TX2
 
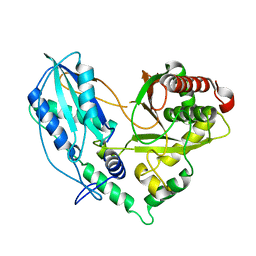 | |
2XHG
 
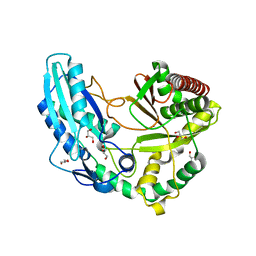 | | Crystal Structure of the Epimerization Domain from the Initiation Module of Tyrocidine Biosynthesis | | Descriptor: | GLYCEROL, SODIUM ION, TYROCIDINE SYNTHETASE A | | Authors: | Samel, S.-A, Heine, A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2010-06-15 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Epimerization Domain of Tyrocidine Synthetase A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HVM
 
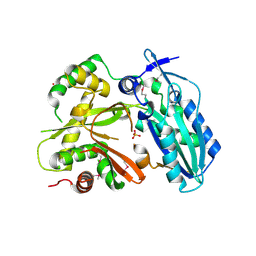 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
4JN5
 
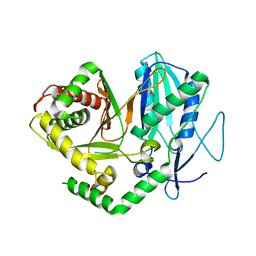 | |
4JN3
 
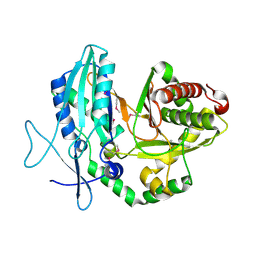 | |
4ZNM
 
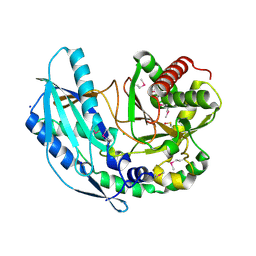 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZXW
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
7JUA
 
 | |
7JTJ
 
 | |
5DIJ
 
 | | The crystal structure of CT | | Descriptor: | CHLORIDE ION, GLYCEROL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5DLK
 
 | | The crystal structure of CT mutant | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-06 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5DUA
 
 | |
5EGF
 
 | | The crystal structure of SeMet-CT | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-10-27 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5DU9
 
 | | First condensation domain of the calcium-dependent antibiotic synthetase in complex with substrate analogue 2a | | Descriptor: | (2S)-2-amino-N-butyl-propanamide, CDA peptide synthetase I, CHLORIDE ION, ... | | Authors: | Bloudoff, K, Alonzo, D.A, Schmeing, T.M. | | Deposit date: | 2015-09-18 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical Probes Allow Structural Insight into the Condensation Reaction of Nonribosomal Peptide Synthetases.
Cell Chem Biol, 23, 2016
|
|
7C1L
 
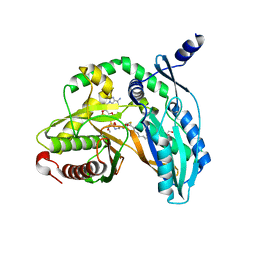 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1U
 
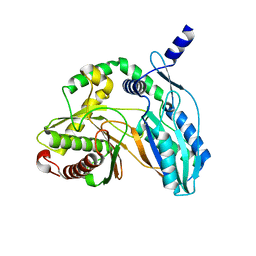 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140V/R148A in a "product-released" conformation | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1R
 
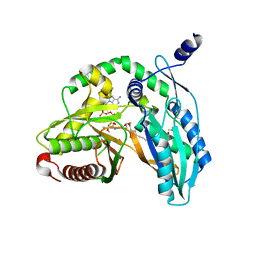 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1H
 
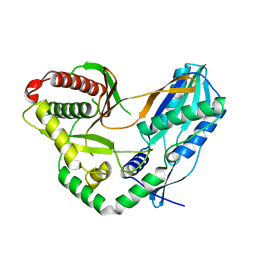 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1P
 
 | | Crystal structure of the starter condensation domain of the rhizomide synthetase RzmA mutant H140V, R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
