4AZJ
 
 | | Structural basis of L-phosphoserine binding to Bacillus alcalophilus phosphoserine aminotransferase | | Descriptor: | CHLORIDE ION, PHOSPHOSERINE, PHOSPHOSERINE AMINOTRANSFERASE, ... | | Authors: | Battula, P, Dubnovitsky, A.P, Papageorgiou, A.C. | | Deposit date: | 2012-06-26 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of L-Phosphoserine Binding to Bacillus Alcalophilus Phosphoserine Aminotransferase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LW4
 
 | |
3KGX
 
 | |
3KE3
 
 | |
2YRR
 
 | | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8 | | Descriptor: | Aminotransferase, class V, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8
To be Published
|
|
3KGW
 
 | |
1BT4
 
 | |
2YRI
 
 | | Crystal structure of alanine-pyruvate aminotransferase with 2-methylserine | | Descriptor: | (S,E)-3-HYDROXY-2-((3-HYDROXY-2-METHYL-5-(PHOSPHONOOXYMETHYL)PYRIDIN-4-YL)METHYLENEAMINO)-2-METHYLPROPANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase, ... | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | hypothetical alanine aminotransferase (TTHA0173) from Thermus thermophilus HB8
To be Published
|
|
3LVM
 
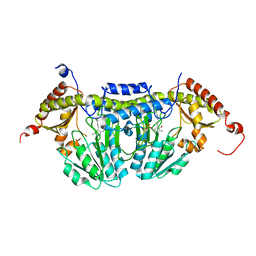 | | Crystal Structure of E.coli IscS | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
2Z9W
 
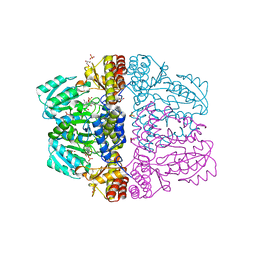 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
1C0N
 
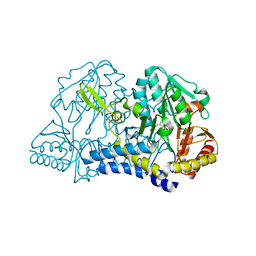 | | CSDB PROTEIN, NIFS HOMOLOGUE | | Descriptor: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | Deposit date: | 1999-07-17 | | Release date: | 2000-07-17 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
2Z9X
 
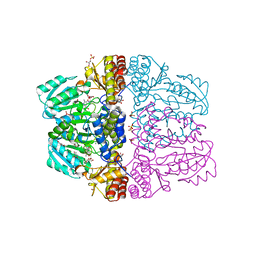 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxyl-L-alanine | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ALANINE, Aspartate aminotransferase, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
1EG5
 
 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-02-13 | | Release date: | 2000-04-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
1ELU
 
 | | COMPLEX BETWEEN THE CYSTINE C-S LYASE C-DES AND ITS REACTION PRODUCT CYSTEINE PERSULFIDE. | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, ... | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1BJN
 
 | |
2YOB
 
 | | High resolution AGXT_M structure | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fabelo-Rosa, I, Mesa-Torres, N, Riverol, D, Yunta, C, Albert, A, Salido, E, Pey, A.L. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Role of Protein Denaturation Energetics and Molecular Chaperones in the Aggregation and Mistargeting of Mutants Causing Primary Hyperoxaluria Type I
Plos One, 8, 2013
|
|
1BJO
 
 | |
2Z9V
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
3M5U
 
 | | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Phosphoserine aminotransferase | | Authors: | Kim, Y, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni
To be Published
|
|
2Z9U
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
1ELQ
 
 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ECX
 
 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kaiser, J.T, Clausen, T.C, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-01-26 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
3A9Z
 
 | |
7XES
 
 | | NifS with L-penicillamine | | Descriptor: | (2~{R})-3-methyl-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-butanoic acid, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XEJ
 
 | | SufS with D-cysteine for 5 min | | Descriptor: | (2~{S})-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-propanoic acid, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamiura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
