3FD9
 
 | |
8GDY
 
 | | Crystal structure of the human PDI first domain with 9 mutations | | Descriptor: | 1,2-ETHANEDIOL, Protein disulfide-isomerase, THIOCYANATE ION | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
8GDU
 
 | | Crystal structure of a mutant methyl transferase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium (NESG) Target MvR53-11M | | Descriptor: | Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Forouhar, F, Banayan, N.E, Loughlin, B.L, Singh, S, Wong, V, Hunt, H.S, Handelman, S.K, Price, N, Hunt, J.F. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Systematic enhancement of protein crystallization efficiency by bulk lysine-to-arginine (KR) substitution.
Protein Sci., 33, 2024
|
|
8GK6
 
 | |
3MWS
 
 | | Crystal Structure of Group N HIV-1 Protease | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | Authors: | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
8BBX
 
 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 in space group C222(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoprotease endo-Pro, TETRAETHYLENE GLYCOL, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
8B57
 
 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-09-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
8DVW
 
 | | Structure of the Campylobacter concisus glycosyltransferase PglA R203Q | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-30 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
8DVZ
 
 | | Structure of the Campylobacter concisus glycosyltransferase PglA R282V variant | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-30 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
8DQD
 
 | | Structure of the Campylobacter concisus glycosyltransferase PglA | | Descriptor: | N, N'-diacetylbacillosaminyl-diphospho-undecaprenol alpha-1,3-N-acetylgalactosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE | | Authors: | Vuksanovic, N, Clasman, J.R, Bernstein, H.M, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-07-18 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Specificity determinants revealed by the structure of glycosyltransferase Campylobacter concisus PglA.
Protein Sci., 33, 2024
|
|
3OEO
 
 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
8CP3
 
 | | Apo-LarE in complex with AMP-PNP | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, NAD_synthase domain-containing protein, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
8CNZ
 
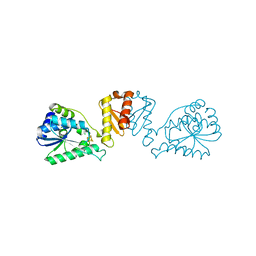 | | mmLarE-[4Fe-4S] phased by Fe-SAD | | Descriptor: | CHLORIDE ION, IODIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Pecqueur, L, Zecchin, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
8CP4
 
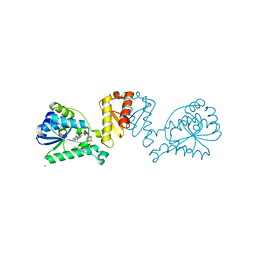 | | [4Fe-4S] cluster containing LarE in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Zecchin, P, Pecqueur, L, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
8PKY
 
 | |
8PM1
 
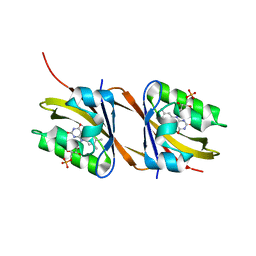 | |
8J4H
 
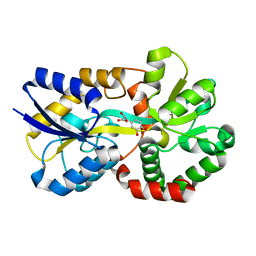 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with Danshensu (DSS) | | Descriptor: | (2~{R})-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-propanoic acid, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
8JLY
 
 | |
8J4J
 
 | | X-ray structure of a ferric ion-binding protein A (FbpA) from Vibrio metschnikovii in complex with ferric ion | | Descriptor: | CARBONATE ION, FE (III) ION, Ferric iron ABC transporter iron-binding protein | | Authors: | Lu, P, Jiang, J, Nagata, K. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism of Fe 3+ binding inhibition to Vibrio metschnikovii ferric ion-binding protein, FbpA, by rosmarinic acid and its hydrolysate, danshensu.
Protein Sci., 33, 2024
|
|
8JJV
 
 | |
8QMF
 
 | | Transketolase from Vibrio vulnificus in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ballut, L, Georges, R.N, Octobre, G, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determination and kinetic analysis of the transketolase from Vibrio vulnificus reveal unexpected cooperative behavior.
Protein Sci., 33, 2024
|
|
8Q8K
 
 | | KI Polyomavirus LTA NLS bound to importin alpha 2 | | Descriptor: | Importin subunit alpha-1, Large T antigen | | Authors: | Cross, E.M, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-08-18 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A functional and structural comparative analysis of large tumor antigens reveals evolution of different importin alpha-dependent nuclear localization signals.
Protein Sci., 33, 2024
|
|
8KB6
 
 | | Crystal Structure of Canine TNF-alpha | | Descriptor: | Tumor necrosis factor | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.850166 Å) | | Cite: | Structure-based development of a canine TNF-alpha-specific antibody using adalimumab as a template.
Protein Sci., 33, 2024
|
|
8OI2
 
 | | Crystal structure of Alb1 megabody in complex with human serum albumin | | Descriptor: | Alb1 Megabody, Albumin | | Authors: | De Felice, S, Zoia, G, Romanyuk, Z, Pardon, E, Steyaert, J, Angelini, A, Cendron, L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of human serum albumin in complex with megabody reveals unique human and murine cross-reactive binding site.
Protein Sci., 33, 2024
|
|
8STT
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
