1KSL
 
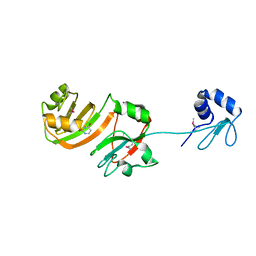 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSM
 
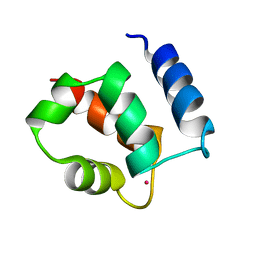 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | Deposit date: | 2002-01-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
1KSN
 
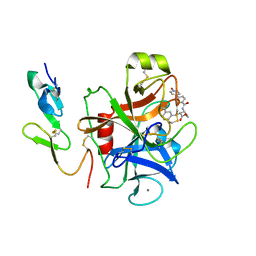 | | Crystal Structure of Human Coagulation Factor XA Complexed with FXV673 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XA, METHYL-3-(4'-N-OXOPYRIDYLPHENOYL)-3-METHYL-2-(M-AMIDINOBENZYL)-PROPIONATE | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-01-14 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of the beta-aminoester class of factor Xa inhibitors. Part 2: Identification of FXV673 as a potent and selective inhibitor with excellent In vivo anticoagulant activity.
Bioorg.Med.Chem.Lett., 12, 2002
|
|
1KSO
 
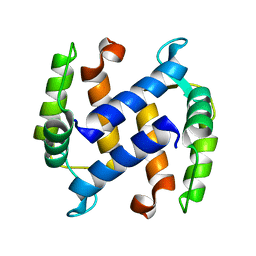 | | CRYSTAL STRUCTURE OF APO S100A3 | | Descriptor: | S100 CALCIUM-BINDING PROTEIN A3 | | Authors: | Mittl, P.R, Fritz, G, Sargent, D.F, Richmond, T.J, Heizmann, C.W, Grutter, M.G. | | Deposit date: | 2002-01-14 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-free MIRAS phasing: structure of apo-S100A3.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KSP
 
 | | DNA polymerase I Klenow fragment (E.C.2.7.7.7) mutant/DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*PST)-3'), PROTEIN (DNA POLYMERASE I-KLENOW FRAGMENT (E.C.2.7.7.7)), ZINC ION | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-19 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
1KSQ
 
 | | NMR Study of the Third TB Domain from Latent Transforming Growth Factor-beta Binding Protein-1 | | Descriptor: | LATENT TRANSFORMING GROWTH FACTOR BETA BINDING PROTEIN 1 | | Authors: | Lack, J, O'leary, J.M, Knott, V, Yuan, X, Rifkin, D.B, Handford, P.A, Downing, A.K. | | Deposit date: | 2002-01-14 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Third TB Domain from LTBP1 Provides Insight into Assembly
of the Large Latent Complex that Sequesters Latent TGF-beta.
J.Mol.Biol., 334, 2003
|
|
1KSR
 
 | | THE REPEATING SEGMENTS OF THE F-ACTIN CROSS-LINKING GELATION FACTOR (ABP-120) HAVE AN IMMUNOGLOBULIN FOLD, NMR, 20 STRUCTURES | | Descriptor: | GELATION FACTOR | | Authors: | Fucini, P, Renner, C, Herberhold, C, Noegel, A.A, Holak, T.A. | | Deposit date: | 1997-02-07 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold.
Nat.Struct.Biol., 4, 1997
|
|
1KSS
 
 | | Crystal Structure of His505Ala Mutant Flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-01-14 | | Release date: | 2002-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina.
Biochemistry, 41, 2002
|
|
1KSU
 
 | | Crystal Structure of His505Tyr Mutant Flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-01-14 | | Release date: | 2002-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina.
Biochemistry, 41, 2002
|
|
1KSV
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URIDINE-5'-MONOPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSW
 
 | | Structure of Human c-Src Tyrosine Kinase (Thr338Gly Mutant) in Complex with N6-benzyl ADP | | Descriptor: | N6-BENZYL ADENOSINE-5'-DIPHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Witucki, L.A, Huang, X, Shah, K, Liu, Y, Kyin, S, Eck, M.J, Shokat, K.M. | | Deposit date: | 2002-01-14 | | Release date: | 2002-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme.
Chem.Biol., 9, 2002
|
|
1KSX
 
 | |
1KSY
 
 | | Crystal Structures of Two Intermediates in the Assembly of the Papillomavirus Replication Initiation Complex | | Descriptor: | E1 Recognition Sequence, Strand 1, Strand 2, ... | | Authors: | Enemark, E.J, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2002-01-14 | | Release date: | 2002-03-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex.
EMBO J., 21, 2002
|
|
1KSZ
 
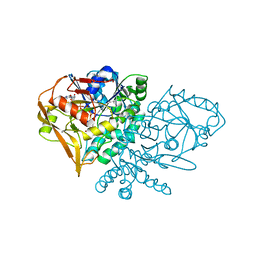 | | ENTRAPMENT OF 6-THIOPHOSPHORYL-IMP IN THE ACTIVE SITE OF CRYSTALLINE ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI, DATA COLLECTED AT 298K | | Descriptor: | 2-DEAZO-6-THIOPHOSPHATE GUANOSINE-5'-MONOPHOSPHATE, ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Poland, B.W, Bruns, C.A, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1997-01-14 | | Release date: | 1997-10-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Entrapment of 6-thiophosphoryl-IMP in the active site of crystalline adenylosuccinate synthetase from Escherichia coli.
J.Biol.Chem., 272, 1997
|
|
1KT0
 
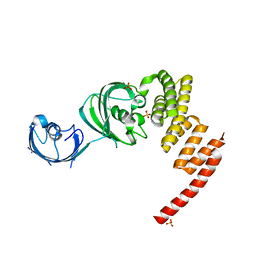 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | 51 KDA FK506-BINDING PROTEIN, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a
component of steroid receptor complexes
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1KT1
 
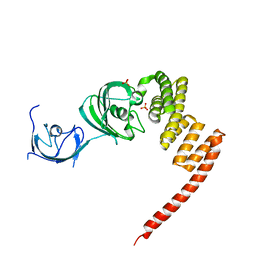 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | FK506-binding protein FKBP51, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STRUCTURE OF THE LARGE FK506-BINDING PROTEIN FKBP51, AN HSP90-BINDING PROTEIN AND A COMPONENT OF STEROID RECEPTOR COMPLEXES
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1KT2
 
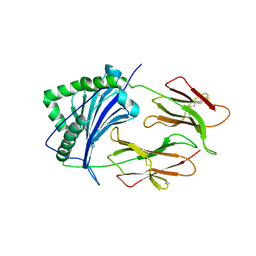 | | CRYSTAL STRUCTURE OF CLASS II MHC MOLECULE IEK BOUND TO MOTH CYTOCHROME C PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion protein consisting of cytochrome C peptide, ... | | Authors: | Fremont, D.H, Dai, S, Chiang, H, Crawford, F, Marrack, P, Kappler, J. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of cytochrome c presentation by IE(k).
J.Exp.Med., 195, 2002
|
|
1KT3
 
 | | Crystal structure of bovine holo-RBP at pH 2.0 | | Descriptor: | Plasma retinol-binding protein, RETINOL | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution Structures of
Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KT4
 
 | | Crystal structure of bovine holo-RBP at pH 3.0 | | Descriptor: | RETINOL, plasma retinol-binding protein | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KT5
 
 | | Crystal structure of bovine holo-RBP at pH 4.0 | | Descriptor: | Plasma retinol-binding protein, RETINOL | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KT6
 
 | | Crystal structure of bovine holo-RBP at pH 9.0 | | Descriptor: | RETINOL, plasma retinol-binding protein | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KT7
 
 | | Crystal structure of bovine holo-RBP at pH 7.0 | | Descriptor: | Plasma retinol-binding protein, RETINOL | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KT8
 
 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL): THREE DIMENSIONAL STRUCTURE OF ENZYME IN ITS KETIMINE FORM WITH THE SUBSTRATE L-ISOLEUCINE | | Descriptor: | ACETIC ACID, BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, MITOCHONDRIAL, ... | | Authors: | Yennawar, N.H, Conway, M.E, Yennawar, H.P, Farber, G.K, Hutson, S.M. | | Deposit date: | 2002-01-15 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human mitochondrial branched chain aminotransferase reaction intermediates: ketimine and pyridoxamine phosphate forms
Biochemistry, 41, 2002
|
|
1KT9
 
 | | Crystal Structure of C. elegans Ap4A Hydrolase | | Descriptor: | Diadenosine Tetraphosphate Hydrolase | | Authors: | Bailey, S, Sedelnikova, S.E, Blackburn, G.M, Abdelghany, H.M, Baker, P.J, McLennan, A.G, Rafferty, J.B. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms
Structure, 10, 2002
|
|
1KTA
 
 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE : THREE DIMENSIONAL STRUCTURE OF THE ENZYME IN ITS PYRIDOXAMINE PHOSPHATE FORM. | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETIC ACID, ... | | Authors: | Yennawar, N.H, Conway, M.E, Yennawar, H.P, Farber, G.K, Hutson, S.M. | | Deposit date: | 2002-01-15 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human mitochondrial branched chain aminotransferase reaction intermediates: ketimine and pyridoxamine phosphate forms
Biochemistry, 41, 2002
|
|
