1K3Y
 
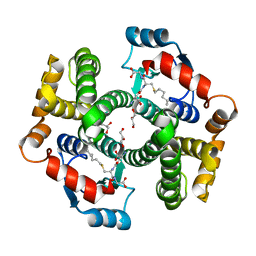 | | Crystal Structure Analysis of human Glutathione S-transferase with S-hexyl glutatione and glycerol at 1.3 Angstrom | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1, GLYCEROL, S-HEXYLGLUTATHIONE | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
1K3Z
 
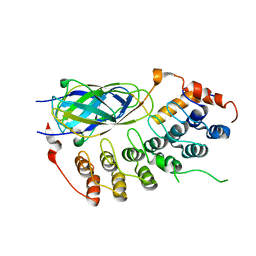 | | X-ray crystal structure of the IkBb/NF-kB p65 homodimer complex | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Shiva, M, Huang, D.B, Chen, Y, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
1K40
 
 | |
1K41
 
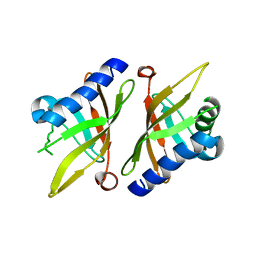 | | Crystal structure of KSI Y57S mutant | | Descriptor: | Ketosteroid Isomerase | | Authors: | Cha, S.S, Oh, B.H, Nam, G.H, Jang, D.S, Lee, T.H, Choi, K.Y. | | Deposit date: | 2001-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Maintenance of alpha-helical structures by phenyl rings in the active-site tyrosine triad contributes to catalysis and stability of ketosteroid isomerase from Pseudomonas putida biotype B
Biochemistry, 40, 2001
|
|
1K42
 
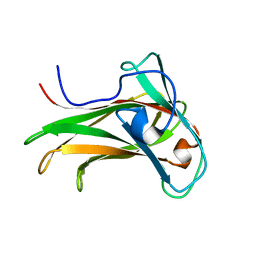 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
1K43
 
 | | 10 Structure Ensemble of the 14-residue peptide RG-KWTY-NG-ITYE-GR (MBH12) | | Descriptor: | MBH12 | | Authors: | Pastor, M.T, Lopez de la Paz, M, Lacroix, E, Serrano, L, Perez-Paya, E. | | Deposit date: | 2001-10-05 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K44
 
 | |
1K45
 
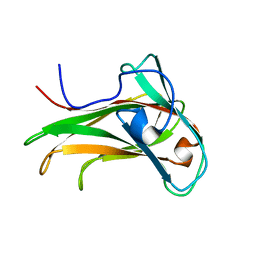 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
1K46
 
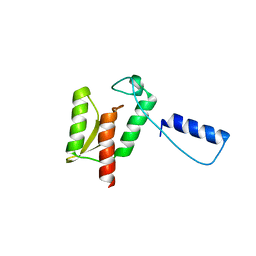 | | Crystal Structure of the Type III Secretory Domain of Yersinia YopH Reveals a Domain-Swapped Dimer | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH | | Authors: | Smith, C.L, Khandelwal, P, Keliikuli, K, Zuiderweg, E.R.P, Saper, M.A. | | Deposit date: | 2001-10-05 | | Release date: | 2001-11-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the type III secretion and substrate-binding domain of Yersinia YopH phosphatase.
Mol.Microbiol., 42, 2001
|
|
1K47
 
 | |
1K48
 
 | |
1K49
 
 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase (cation free form) | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, SULFATE ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-06 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
1K4A
 
 | | STRUCTURE OF AGAA RNA TETRALOOP, NMR, 20 STRUCTURES | | Descriptor: | 5'-R(*GP*GP*UP*UP*CP*AP*GP*AP*AP*GP*AP*AP*CP*C)-3' | | Authors: | Wu, H, Yang, P.K, Butcher, S.E, Kang, S, Chanfreau, G, Feigon, J. | | Deposit date: | 2001-10-07 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.
EMBO J., 20, 2001
|
|
1K4B
 
 | | STRUCTURE OF AGUU RNA TETRALOOP, NMR, 20 STRUCTURES | | Descriptor: | 5'-R(*GP*GP*UP*UP*CP*AP*GP*UP*UP*GP*AP*AP*CP*C)-3' | | Authors: | Wu, H, Yang, P.K, Butcher, S.E, Kang, S, Chanfreau, G, Feigon, J. | | Deposit date: | 2001-10-07 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.
EMBO J., 20, 2001
|
|
1K4C
 
 | | Potassium Channel KcsA-Fab complex in high concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
1K4D
 
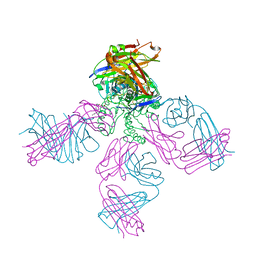 | | Potassium Channel KcsA-Fab complex in low concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
1K4E
 
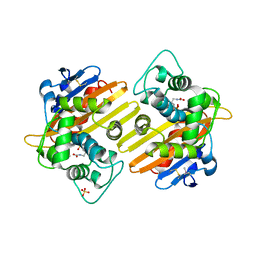 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASES OXA-10 DETERMINED BY MAD PHASING WITH SELENOMETHIONINE | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURE OF CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K4F
 
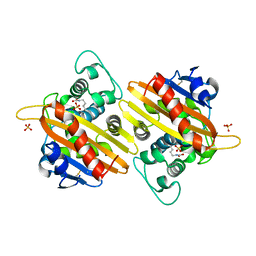 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 AT 1.6 A RESOLUTION | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the class D beta-lactamase OXA-2
To be Published
|
|
1K4G
 
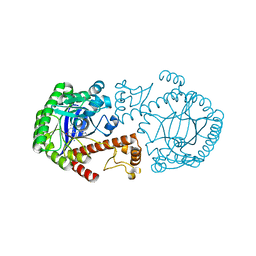 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
1K4H
 
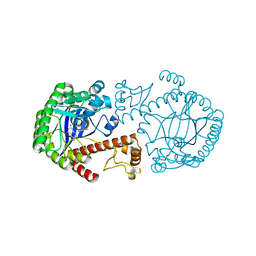 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-Diamino-8-propylsulfanylmethyl-3H-quinazoline-4-one | | Descriptor: | 2,6-DIAMINO-8-PROPYLSULFANYLMETHYL-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE-TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
Chembiochem, 3, 2002
|
|
1K4I
 
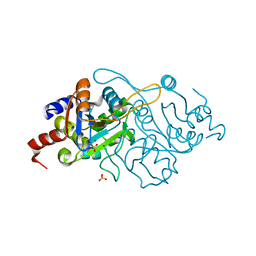 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with two Magnesium ions | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, MAGNESIUM ION, SULFATE ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
1K4J
 
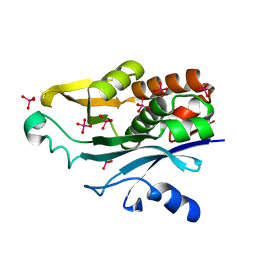 | | Crystal Structure of the Acyl-homoserinelactone Synthase EsaI Complexed with Rhenate | | Descriptor: | PERRHENATE, acyl-homoserinelactone synthase EsaI | | Authors: | Watson, W.T, Minogue, T.D, Val, D.L, Beck von Bodman, S, Churchill, M.E.A. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing.
Mol.Cell, 9, 2002
|
|
1K4K
 
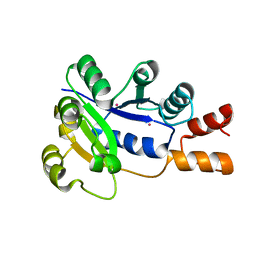 | | Crystal structure of E. coli Nicotinic acid mononucleotide adenylyltransferase | | Descriptor: | Nicotinic acid mononucleotide adenylyltransferase, XENON | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A.L. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
1K4L
 
 | | Crystal Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase in complex with two Manganese ions | | Descriptor: | 3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase, MANGANESE (II) ION, SULFATE ION | | Authors: | Liao, D.-I, Zheng, Y.-J, Viitanen, P.V, Jordan, D.B. | | Deposit date: | 2001-10-08 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural definition of the active site and catalytic mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase.
Biochemistry, 41, 2002
|
|
1K4M
 
 | | Crystal structure of E.coli nicotinic acid mononucleotide adenylyltransferase complexed to deamido-NAD | | Descriptor: | CITRIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NaMN adenylyltransferase | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
