1B16
 
 | | ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS TERNARY COMPLEX WITH NAD-3-PENTANONE | | Descriptor: | NICOTINAMIDE ADENINE DINUCLEOTIDE 3-PENTANONE ADDUCT, PROTEIN (ALCOHOL DEHYDROGENASE) | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-25 | | Release date: | 1999-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
1B17
 
 | |
1B18
 
 | |
1B19
 
 | |
1B1A
 
 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Hoffmann, B, Konrat, R, Bothe, H, Buckel, W, Kraeutler, B. | | Deposit date: | 1998-11-19 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium.
Eur.J.Biochem., 263, 1999
|
|
1B1B
 
 | | IRON DEPENDENT REGULATOR | | Descriptor: | PROTEIN (IRON DEPENDENT REGULATOR), SULFATE ION, ZINC ION | | Authors: | Pohl, E, Holmes, R.K, Hol, W.G. | | Deposit date: | 1998-11-19 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied.
J.Mol.Biol., 285, 1999
|
|
1B1C
 
 | | CRYSTAL STRUCTURE OF THE FMN-BINDING DOMAIN OF HUMAN CYTOCHROME P450 REDUCTASE AT 1.93A RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH-CYTOCHROME P450 REDUCTASE) | | Authors: | Zhao, Q, Modi, S, Smith, G, Paine, M, Mcdonagh, P.D, Wolf, C.R, Tew, D, Lian, L.-Y, Roberts, G.C.K, Driessen, H.P.C. | | Deposit date: | 1998-11-19 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Protein Sci., 8, 1999
|
|
1B1E
 
 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN VARIANT K40Q | | Descriptor: | CITRIC ACID, HYDROLASE ANGIOGENIN | | Authors: | Leonidas, D.D, Acharya, K.R. | | Deposit date: | 1998-11-20 | | Release date: | 1999-04-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structures of native human angiogenin and two active site variants: implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth.
J.Mol.Biol., 285, 1999
|
|
1B1G
 
 | |
1B1H
 
 | | OLIGO-PEPTIDE BINDING PROTEIN/TRIPEPTIDE (LYS HPE LYS) COMPLEX | | Descriptor: | PROTEIN (LYS HPE LYS), PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), URANYL (VI) ION | | Authors: | Davies, T.G, Tame, J.R.H. | | Deposit date: | 1998-11-10 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relating structure to thermodynamics: the crystal structures and binding affinity of eight OppA-peptide complexes.
Protein Sci., 8, 1999
|
|
1B1I
 
 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN | | Descriptor: | CITRIC ACID, HYDROLASE ANGIOGENIN | | Authors: | Leonidas, D.D, Acharya, K.R. | | Deposit date: | 1998-11-20 | | Release date: | 1999-04-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structures of native human angiogenin and two active site variants: implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth.
J.Mol.Biol., 285, 1999
|
|
1B1J
 
 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN VARIANT H13A. | | Descriptor: | HYDROLASE ANGIOGENIN | | Authors: | Leonidas, D.D, Acharya, K.R. | | Deposit date: | 1998-11-20 | | Release date: | 1999-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structures of native human angiogenin and two active site variants: implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth.
J.Mol.Biol., 285, 1999
|
|
1B1U
 
 | |
1B1V
 
 | | NMR STRUCTURE OF PSP1, PLASMATOCYTE-SPREADING PEPTIDE FROM PSEUDOPLUSIA INCLUDENS | | Descriptor: | PROTEIN (PLASMATOCYTE-SPREADING PEPTIDE) | | Authors: | Volkman, B.F, Clark, K.D, Anderson, M.E, Pech, L.L, Markley, J.L, Strand, M.R. | | Deposit date: | 1998-11-23 | | Release date: | 1998-12-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the insect cytokine peptide plasmatocyte-spreading peptide 1 from Pseudoplusia includens.
J.Biol.Chem., 274, 1999
|
|
1B1X
 
 | |
1B1Y
 
 | | SEVENFOLD MUTANT OF BARLEY BETA-AMYLASE | | Descriptor: | PROTEIN (BETA-AMYLASE), alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose | | Authors: | Mikami, B, Yoon, H.J, Yoshigi, N. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the sevenfold mutant of barley beta-amylase with increased thermostability at 2.5 A resolution.
J.Mol.Biol., 285, 1999
|
|
1B1Z
 
 | | STREPTOCOCCAL PYROGENIC EXOTOXIN A1 | | Descriptor: | PROTEIN (TOXIN) | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1998-11-24 | | Release date: | 1999-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for the recognition of superantigen streptococcal pyrogenic exotoxin A (SpeA1) by MHC class II molecules and T-cell receptors.
EMBO J., 18, 1999
|
|
1B20
 
 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B21
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B22
 
 | | RAD51 (N-TERMINAL DOMAIN) | | Descriptor: | DNA REPAIR PROTEIN RAD51 | | Authors: | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
1B23
 
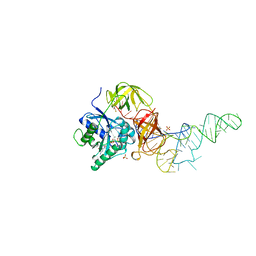 | | E. coli cysteinyl-tRNA and T. aquaticus elongation factor EF-TU:GTP ternary complex | | Descriptor: | CYSTEINE, CYSTEINYL TRNA, ELONGATION FACTOR TU, ... | | Authors: | Nissen, P, Kjeldgaard, M, Thirup, S, Nyborg, J. | | Deposit date: | 1998-12-04 | | Release date: | 1998-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA.
Structure Fold.Des., 7, 1999
|
|
1B24
 
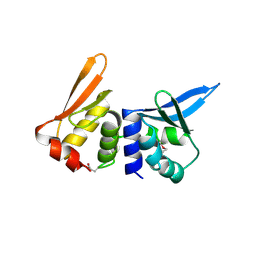 | | I-DMOI, INTRON-ENCODED ENDONUCLEASE | | Descriptor: | Homing endonuclease I-DmoI | | Authors: | Van Roey, P, Silva, G.H. | | Deposit date: | 1998-12-03 | | Release date: | 1999-03-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI.
J.Mol.Biol., 286, 1999
|
|
1B25
 
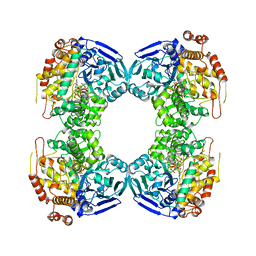 | | FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE), TUNGSTOPTERIN | | Authors: | Hu, Y.L, Faham, S, Roy, R, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1998-12-04 | | Release date: | 1999-03-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 A resolution crystal structure and its mechanistic implications.
J.Mol.Biol., 286, 1999
|
|
1B26
 
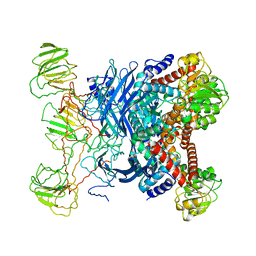 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
1B27
 
 | |
