1ALI
 
 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1ALJ
 
 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1ALK
 
 | |
1ALL
 
 | | ALLOPHYCOCYANIN | | Descriptor: | ALLOPHYCOCYANIN, PHYCOCYANOBILIN | | Authors: | Brejc, K, Ficner, R, Huber, R, Steinbacher, S. | | Deposit date: | 1995-03-01 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Isolation, crystallization, crystal structure analysis and refinement of allophycocyanin from the cyanobacterium Spirulina platensis at 2.3 A resolution.
J.Mol.Biol., 249, 1995
|
|
1ALN
 
 | |
1ALQ
 
 | |
1ALU
 
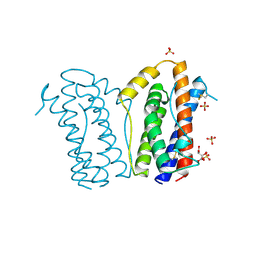 | | HUMAN INTERLEUKIN-6 | | Descriptor: | INTERLEUKIN-6, L(+)-TARTARIC ACID, SULFATE ION | | Authors: | Somers, W.S, Stahl, M, Seehra, J.S. | | Deposit date: | 1997-06-03 | | Release date: | 1998-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling.
EMBO J., 16, 1997
|
|
1ALV
 
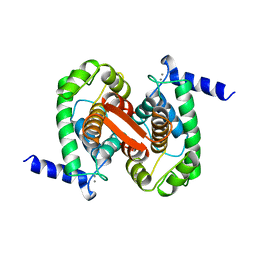 | | CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G, Chattopadhyay, D, Maki, M. | | Deposit date: | 1997-06-03 | | Release date: | 1998-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
1ALW
 
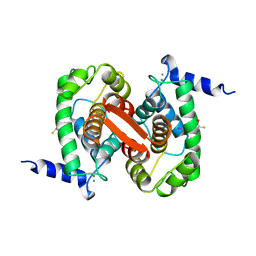 | | INHIBITOR AND CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G. | | Deposit date: | 1997-06-04 | | Release date: | 1998-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
1ALX
 
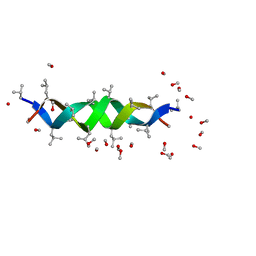 | | GRAMICIDIN D FROM BACILLUS BREVIS (METHANOL SOLVATE) | | Descriptor: | GRAMICIDIN A, METHANOL | | Authors: | Burkhart, B.M, Langs, D.A, Smith, G.D, Courseille, C, Precigoux, G, Hospital, M, Pangborn, W.A, Duax, W.L. | | Deposit date: | 1997-06-05 | | Release date: | 1998-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
1ALY
 
 | |
1ALZ
 
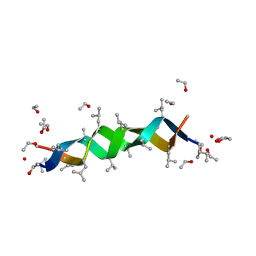 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | ETHANOL, ILE-GRAMICIDIN C, VAL-GRAMICIDIN A | | Authors: | Burkhart, B.M, Pangborn, W.A, Duax, W.L, Langs, D.A. | | Deposit date: | 1997-06-06 | | Release date: | 1998-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
1AM0
 
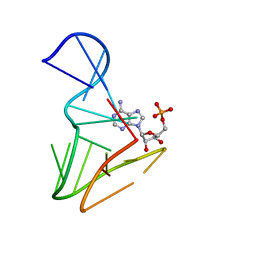 | | AMP RNA APTAMER COMPLEX, NMR, 8 STRUCTURES | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA APTAMER | | Authors: | Jiang, F, Kumar, R.A, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-06-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of RNA Folding and Recognition in an AMP-RNA Aptamer Complex
Nature, 382, 1996
|
|
1AM1
 
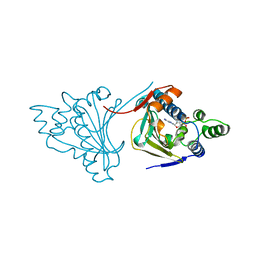 | |
1AM2
 
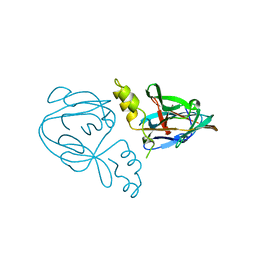 | |
1AM4
 
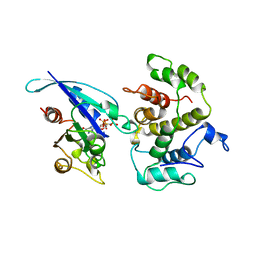 | | COMPLEX BETWEEN CDC42HS.GMPPNP AND P50 RHOGAP (H. SAPIENS) | | Descriptor: | CDC42HS, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P, Gamblin, S.J, Smerdon, S.J. | | Deposit date: | 1997-06-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP.
Nature, 388, 1997
|
|
1AM5
 
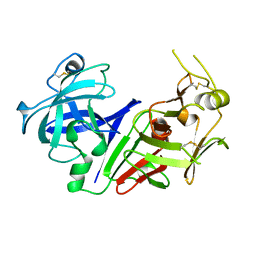 | |
1AM6
 
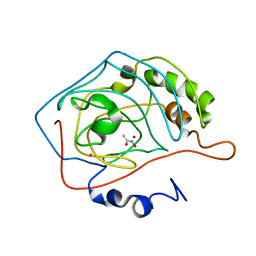 | | CARBONIC ANHYDRASE II INHIBITOR: ACETOHYDROXAMATE | | Descriptor: | ACETOHYDROXAMIC ACID, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Scolnick, L.R, Clements, A.M, Christianson, D.W. | | Deposit date: | 1997-06-24 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Binding Mode of Hydroxamate Inhibitors to Human Carbonic Anhydrase II
J.Am.Chem.Soc., 119, 1997
|
|
1AM7
 
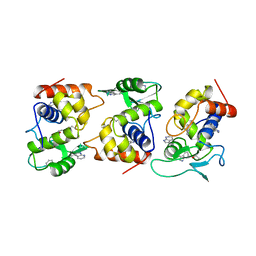 | | Lysozyme from bacteriophage lambda | | Descriptor: | ISOPROPYL ALCOHOL, LYSOZYME | | Authors: | Evrard, C, Fastrez, J, Declercq, J.P. | | Deposit date: | 1997-06-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes.
J.Mol.Biol., 276, 1998
|
|
1AM9
 
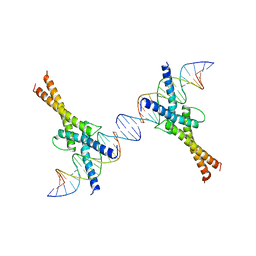 | | HUMAN SREBP-1A BOUND TO LDL RECEPTOR PROMOTER | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*AP*TP*CP*AP*CP*CP*CP*CP*AP*CP*T P*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*GP*TP*GP*GP*GP*GP*TP*GP*AP*TP*CP*T )-3'), MAGNESIUM ION, ... | | Authors: | Parraga, A, Burley, S.K. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution.
Structure, 6, 1998
|
|
1AMA
 
 | |
1AMB
 
 | |
1AMC
 
 | |
1AMD
 
 | |
1AME
 
 | |
