1NWQ
 
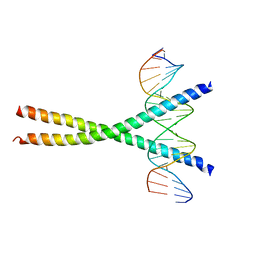 | | CRYSTAL STRUCTURE OF C/EBPALPHA-DNA COMPLEX | | Descriptor: | 5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*TP*GP*CP*GP*CP*AP*AP*TP*AP*GP*GP*A)-3', 5'-D(*TP*TP*CP*CP*TP*AP*TP*TP*GP*CP*GP*CP*AP*AP*TP*CP*CP*AP*GP*TP*T)-3', CCAAT/enhancer binding protein alpha | | Authors: | Miller, M, Shuman, J.D, Sebastian, T, Dauter, Z, Johnson, P.F. | | Deposit date: | 2003-02-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for DNA Recognition by the Basic Region Leucine Zipper
Transcription Factor CCAAT/enhancer Binding Protein Alpha
J.Biol.Chem., 278, 2003
|
|
8K8C
 
 | | Crystal structure of C/EBPalpha BZIP domain bound to a high affinity DNA | | Descriptor: | CCAAT/enhancer-binding protein alpha, DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*A)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*CP*GP*TP*AP*AP*TP*GP*T)-3'), ... | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
6DC0
 
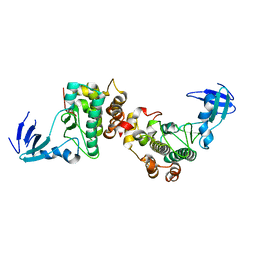 | |
4QL1
 
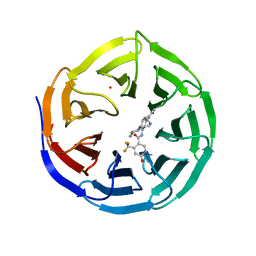 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
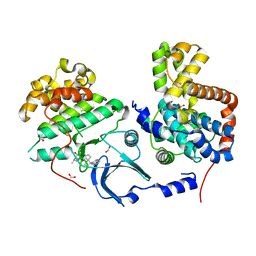
4CRL
 
 | |
6Q6F
 
 | | Crystal structure of IDH1 R132H in complex with HMS101 | | Descriptor: | (2~{R})-2-[2-[(3~{R})-3-(4-fluorophenyl)pyrrolidin-1-yl]ethyl]-1,4-dimethyl-piperazine, Isocitrate dehydrogenase [NADP] cytoplasmic | | Authors: | Chaturvedi, A, Goparaju, R, Gupta, C, Kluenemann, T, Araujo Cruz, M.M, Kloos, A, Goerlich, K, Schottmann, R, Struys, E.A, Ganser, A, Preller, M, Heuser, M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site.
Leukemia, 34, 2020
|
|
