8QWL
 
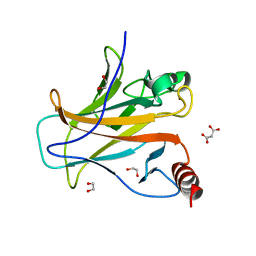 | | Structure of p53 cancer mutant Y163C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, MALONATE ION, ... | | Authors: | Balourdas, D.I, Markl, A.M, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8RQ6
 
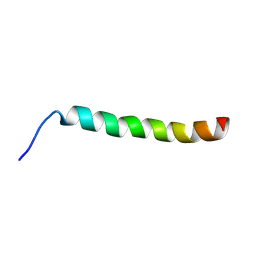 | |
8RF4
 
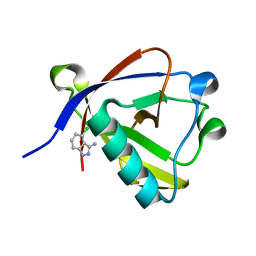 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 9D4 refined against the anomalous diffraction data | | Descriptor: | 4-chloranyl-1~{H}-indazol-3-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RPL
 
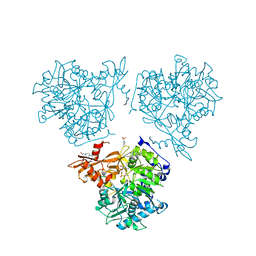 | | AMP-forming acetyl-CoA synthetase from Chloroflexota bacterium with bound acetyl AMP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Acetate--CoA ligase, MAGNESIUM ION, ... | | Authors: | Striska, K, Palm, G.J, Lammers, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Acetyl-CoA synthetase activity is enzymatically regulated by lysine acetylation using acetyl-CoA or acetyl-phosphate as donor molecule.
Nat Commun, 15, 2024
|
|
8QJ3
 
 | |
8QU1
 
 | | mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 1 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Valentin Gese, G, Cipullo, M, Rorbach, J, Hallberg, B.M. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GTPBP8 plays a role in mitoribosome formation in human mitochondria.
Nat Commun, 15, 2024
|
|
1C9Y
 
 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CATALYTIC MECHANISM | | Descriptor: | NORVALINE, ORNITHINE CARBAMOYLTRANSFERASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Yu, X, Morizono, H, Tuchman, M, Allewell, N.M. | | Deposit date: | 1999-08-03 | | Release date: | 2000-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
Proteins, 39, 2000
|
|
1BP7
 
 | | GROUP I MOBILE INTRON ENDONUCLEASE I-CREI COMPLEXED WITH HOMING SITE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP* GP*C)-3'), DNA (5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP* CP*G)-3'), ... | | Authors: | Jurica, M.S, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1998-08-13 | | Release date: | 1999-01-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI.
Mol.Cell, 2, 1998
|
|
1C96
 
 | | S642A:CITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1BR9
 
 | | HUMAN TISSUE INHIBITOR OF METALLOPROTEINASE-2 | | Descriptor: | METALLOPROTEINASE-2 INHIBITOR | | Authors: | Tuuttila, A, Morgunova, E, Bergmann, U, Lindqvist, Y, Tryggvason, K, Schneider, G. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of human tissue inhibitor of metalloproteinases-2 at 2.1 A resolution.
J.Mol.Biol., 284, 1998
|
|
7UHW
 
 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 120 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UHX
 
 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 150 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UDD
 
 | | Horse liver alcohol dehydrogenase complexed with NAD+ and 2,2,2-trifluoroethanol at 150 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-18 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UHV
 
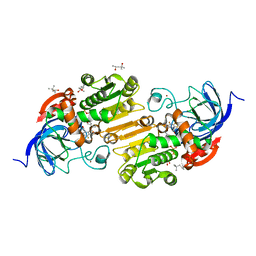 | | Horse liver alcohol dehydrogenase G173A, complexed with NAD+ and 2,3,4,5,6-pentafluorobenzyl alcohol at 50 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
1CA6
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
1CCJ
 
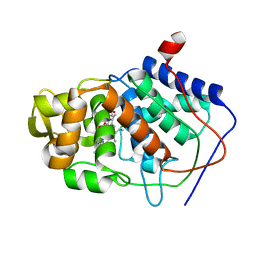 | | CONFORMER SELECTION BY LIGAND BINDING OBSERVED WITH PROTEIN CRYSTALLOGRAPHY | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein conformer selection by ligand binding observed with crystallography.
Protein Sci., 7, 1998
|
|
1BS3
 
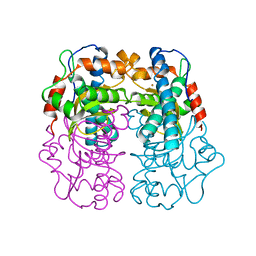 | | P.SHERMANII SOD(FE+3) FLUORIDE | | Descriptor: | FE (III) ION, FLUORIDE ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-08-31 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
1CBO
 
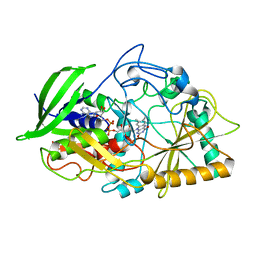 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES HIS447ASN MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1999-02-26 | | Release date: | 1999-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
1BSM
 
 | | P.SHERMANII SOD(FE+3) 140K PH8 | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-08-28 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
5EGH
 
 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase in complex with phosphocholine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
1BMT
 
 | | HOW A PROTEIN BINDS B12: A 3.O ANGSTROM X-RAY STRUCTURE OF THE B12-BINDING DOMAINS OF METHIONINE SYNTHASE | | Descriptor: | CO-METHYLCOBALAMIN, METHIONINE SYNTHASE | | Authors: | Drennan, C.L, Huang, S, Drummond, J.T, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1994-09-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase.
Science, 266, 1994
|
|
1BJZ
 
 | |
7BM4
 
 | | Crystal structure of alpha Carbonic anhydrase from Schistosoma mansoni bound to 1-(4-fluorophenyl)-3-(4-sulphamoylphenyl)selenourea | | Descriptor: | 1-(4-fluorophenyl)-3-(4-sulfamoylphenyl)selenourea, 2-acetamido-2-deoxy-beta-D-glucopyranose, Carbonic anhydrase, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into Schistosoma mansoni Carbonic Anhydrase (SmCA) Inhibition by Selenoureido-Substituted Benzenesulfonamides.
J.Med.Chem., 64, 2021
|
|
1BT8
 
 | | P.SHERMANII SOD(FE+3) PH 10.0 | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
8DL2
 
 | | BoGH13ASus from Bacteroides ovatus bound to acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
