6CVL
 
 | | Crystal structure of the Escherichia coli ATPgS-bound MetNI methionine ABC transporter in complex with its MetQ binding protein | | Descriptor: | IODIDE ION, MERCURY (II) ION, MetI transmembrane subunit, ... | | Authors: | Nguyen, P.T, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Noncanonical role for the binding protein in substrate uptake by the MetNI methionine ATP Binding Cassette (ABC) transporter.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7MRK
 
 | | Chicken CNTN4 APP complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4, DI(HYDROXYETHYL)ETHER | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7B58
 
 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Cl determined at 1.72 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7RMX
 
 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MRN
 
 | | Mouse CNTN5 APP complex | | Descriptor: | Contactin-5, N-APP | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
7AMH
 
 | | SmBRD3(2), Second Bromodomain of Bromodomain 3 from Schistosoma mansoni in complex with DM-A-33, an iBET726 analogue | | Descriptor: | 4-{(2S, 4R)-1-acetyl-4-[(1-benzothiophen-6-yl)amino]-2-methyl-1,2,3,4-tetrahydroquinolin-6-yl}benzoic acid, Putative bromodomain-containing protein 3, ... | | Authors: | McArdle, D, Schiedel, M, McDonough, M.A, Conway, S.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | SBM3_2, Second Bromodomain of Bromodomain 3 from Schistosoma mansoni in complex with DM1, an iBET726 analogue
To Be Published
|
|
8AIW
 
 | | Structure of the K5/CagI complex | | Descriptor: | Cag pathogenicity island protein (Cag19), Designed Ankyrin Repeat Protein K5 | | Authors: | Blanc, M, Guerin, J, Terradot, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed Ankyrin Repeat Proteins provide insights into the structure and function of CagI and are potent inhibitors of CagA translocation by the Helicobacter pylori type IV secretion system.
Plos Pathog., 19, 2023
|
|
8TQ0
 
 | |
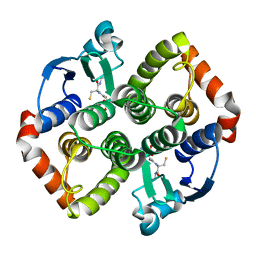
8AI9
 
 | |
8TSU
 
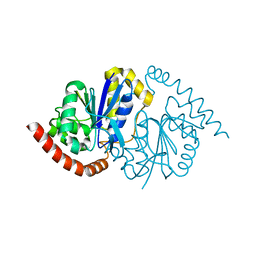 | |
7B59
 
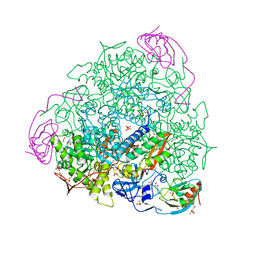 | | X-ray crystal structure of Sporosarcina pasteurii urease inhibited by Ag(PEt3)Br determined at 1.63 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetic and structural analysis of the inactivation of urease by mixed-ligand phosphine halide Ag(I) complexes.
J.Inorg.Biochem., 218, 2021
|
|
7RNQ
 
 | |
6CB2
 
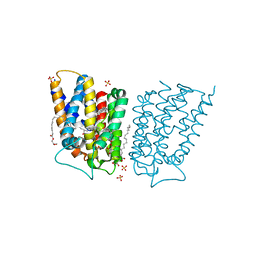 | | Crystal structure of Escherichia coli UppP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Undecaprenyl-diphosphatase | | Authors: | Workman, S.D, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling.
Nat Commun, 9, 2018
|
|
7ROF
 
 | |
8TSX
 
 | |
7AQX
 
 | |
7RK7
 
 | | The complex between TIL 1383i TCR and human Class I MHC HLA-A2 with the bound Tyrosinase(369-377)(N371D) nonameric peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Singh, N.K, Davancaze, L.M, Arbuiso, A, Weiss, L.I, Keller, G.L.J, Baker, B.M. | | Deposit date: | 2021-07-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The complex between TIL 1383i TCR and human Class I MHC HLA-A2 with the bound Tyrosinase(369-377)(N371D) nonameric peptide
To Be Published
|
|
8B05
 
 | | TRYPTOPHAN SYNTHASE - Cryo-trapping by the spitrobot crystal plunger after 20 sec | | Descriptor: | CESIUM ION, CHLORIDE ION, INDOLE, ... | | Authors: | Mehrabi, P, Sung, S, von Stetten, D, Prester, A, Hatton, C.E, Kleine-Dopke, S, Berkes, A, Gore, G, Leimkohl, J.P, Schikora, H, Kollewe, M, Rohde, H, Wilmanns, M, Tellkamp, F, Schulz, E.C. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography.
Nat Commun, 14, 2023
|
|
7AYX
 
 | |
7QXK
 
 | | As isolated MSOX movie series dataset 1 (0.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QY5
 
 | | Crystal structure of the S.pombe Ars2-Red1 complex. | | Descriptor: | NURS complex subunit pir2, RNA elimination defective protein Red1, ZINC ION | | Authors: | Foucher, A.E, Kadlec, J. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
6CED
 
 | | Crystal structure of fragment 3-(3-Methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
8TT1
 
 | |
7R0X
 
 | |
7R3V
 
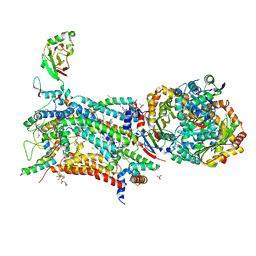 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor CK-2-67. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Pinthong, N, Amporndanai, K, O'Neill, P.M, Hasnain, S.S, Antonyuk, S. | | Deposit date: | 2022-02-07 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting the Ubiquinol-Reduction (Q i ) Site of the Mitochondrial Cytochrome bc 1 Complex for the Development of Next Generation Quinolone Antimalarials.
Biology (Basel), 11, 2022
|
|
