8RPG
 
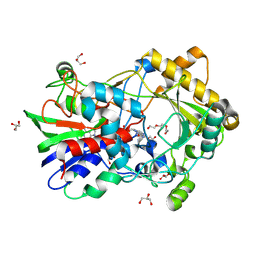 | | Crystal structure of an alcohol oxidase from Streptomyces hiroshimensis | | Descriptor: | Alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Ferreira, P, Cinca-Fernando, P. | | Deposit date: | 2024-01-15 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery, characterization, and synthetic potential of two novel bacterial aryl-alcohol oxidases.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
8PE1
 
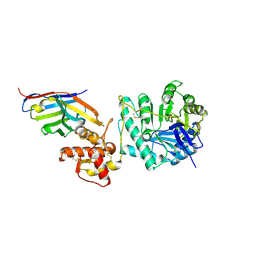 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PE2
 
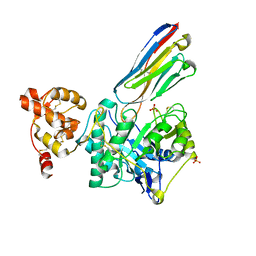 | | Crystal structure of Gel4 in complex with Nanobody 3 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 3, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3GQ1
 
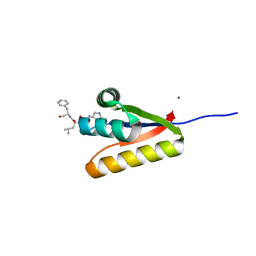 | | The structure of the caulobacter crescentus clpS protease adaptor protein in complex with a WLFVQRDSKE decapeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, WLFVQRDSKE peptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GW1
 
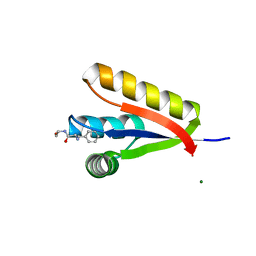 | | The structure of the Caulobacter crescentus CLPs protease adaptor protein in complex with FGG tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS, FGG peptide, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G19
 
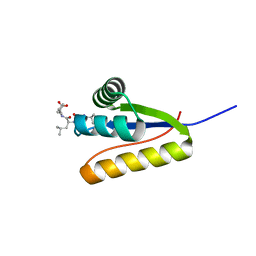 | | The structure of the Caulobacter crescentus clpS protease adaptor protein in complex with LLL tripeptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, LLL tripeptide | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GQ0
 
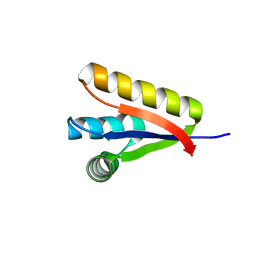 | |
3G1B
 
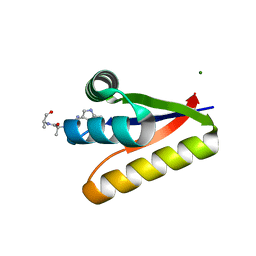 | | The structure of the M53A mutant of Caulobacter crescentus clpS protease adaptor protein in complex with WLFVQRDSKE peptide | | Descriptor: | 10-residue peptide, ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION | | Authors: | Baker, T.A, Roman-Hernandez, G, Sauer, R.T, Grant, R.A. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Molecular basis of substrate selection by the N-end rule adaptor protein ClpS.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G3P
 
 | |
1R9V
 
 | |
7SAM
 
 | |
8CLR
 
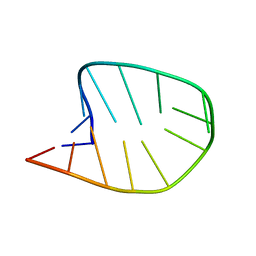 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | Descriptor: | RNA hairpin with CUUG tetraloop | | Authors: | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
6SSO
 
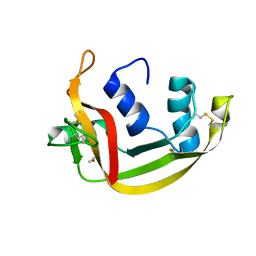 | | EDN mutant L45H | | Descriptor: | ACETATE ION, Non-secretory ribonuclease | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and functional characterization of new family enzymes derivates from human RNase 1 and 3 with antimicrobial and ribonuclease activity
To Be Published
|
|
7RIX
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIW
 
 | | RNA polymerase II elongation complex scaffold 2, without polyamide | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIM
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIY
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 soaked with UTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIP
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 soaked with CTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RIQ
 
 | | RNA polymerase II elongation complex scaffold 1 without polyamide | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6VAR
 
 | | 61 nt human Hepatitis B virus epsilon pre-genomic RNA | | Descriptor: | RNA (61-MER) | | Authors: | LeBlanc, R.M, Kasprzak, W.K, Longhini, A.P, Abulwerdi, F, Ginocchio, S, Shields, B, Nyman, J, Svirydava, M, Del Vecchio, C, Ivanic, J, Schneekloth, J.S, Dayie, T.K, Shapiro, B.A, Le Grice, S.F.J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural insights of the conserved "priming loop" of hepatitis B virus pre-genomic RNA.
J.Biomol.Struct.Dyn., 2021
|
|
4V74
 
 | | 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre5b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8PFK
 
 | | RNA structure with 1-methylpseudouridine, C2 space group | | Descriptor: | MAGNESIUM ION, RNA (12-mer) | | Authors: | Spingler, B, McAuley, K, Nievergelt, P, Thorn, A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-01-31 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | RNA oligomers at atomic resolution containing 1-methylpseudouridine, an essential building block of mRNA vaccines.
Chemmedchem, 19, 2024
|
|
4V83
 
 | | Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V84
 
 | | Crystal structure of a complex containing domain 3 of CrPV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
9MTY
 
 | |
