3U6N
 
 | | Open Structure of the BK channel Gating Ring | | Descriptor: | CALCIUM ION, High-Conductance Ca2+-Activated K+ Channel protein | | Authors: | Yuan, P, MacKinnon, R. | | Deposit date: | 2011-10-12 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Open structure of the Ca(2+) gating ring in the high-conductance Ca(2+)-activated K(+) channel.
Nature, 481, 2011
|
|
1DJR
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER COMPLEXED WITH M-CARBOXYPHENYL-ALPHA-D-GALACTOSE | | Descriptor: | BENZOIC ACID, GLYCEROL, HEAT-LABILE ENTEROTOXIN, ... | | Authors: | Minke, W.E, Pickens, J, Merritt, E.A, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 1999-12-03 | | Release date: | 2000-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of m-carboxyphenyl-alpha-D-galactopyranoside complexed to heat-labile enterotoxin at 1.3 A resolution: surprising variations in ligand-binding modes.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7M5E
 
 | |
3GM8
 
 | | Crystal structure of a beta-glycosidase from Bacteroides vulgatus | | Descriptor: | GLYCEROL, Glycoside hydrolase family 2, candidate beta-glycosidase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a beta-glycosidase from Bacteroides vulgatus
To be Published
|
|
3GVB
 
 | |
7LQL
 
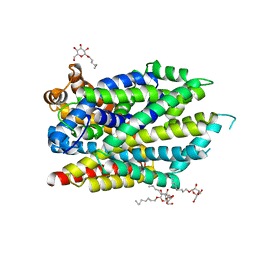 | | Crystal structure of the R375D mutant of LeuT | | Descriptor: | ALANINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Font, J, Aguilar, J, Galli, A, Ryan, R. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Psychomotor impairments and therapeutic implications revealed by a mutation associated with infantile Parkinsonism-Dystonia.
Elife, 10, 2021
|
|
5WVG
 
 | |
3U2D
 
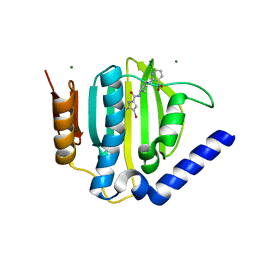 | | S. aureus GyrB ATPase domain in complex with small molecule inhibitor | | Descriptor: | 4-bromo-5-methyl-N-[1-(3-nitropyridin-2-yl)piperidin-4-yl]-1H-pyrrole-2-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
3GO0
 
 | |
3GO5
 
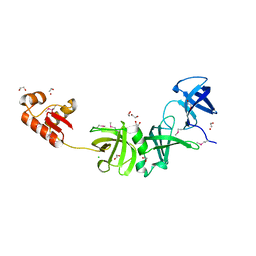 | |
4URT
 
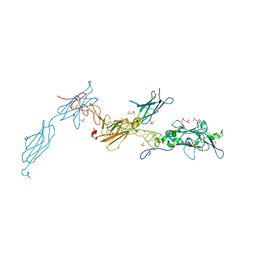 | | The crystal structure of a fragment of netrin-1 in complex with FN5- FN6 of DCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Finci, L.I, Krueger, N, Sun, X, Zhang, J, Chegkazi, M, Wu, Y, Schenk, G, Mertens, H.D.T, Svergun, D.I, Zhang, Y, Wang, J.-h, Meijers, R. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structure of Netrin-1 in Complex with Dcc Reveals the Bi-Functionality of Netrin-1 as a Guidance Cue
Neuron, 83, 2014
|
|
3GOH
 
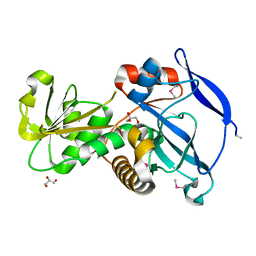 | |
3GOR
 
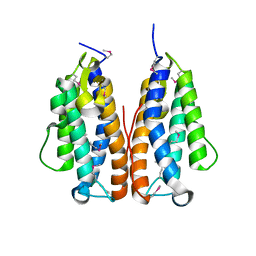 | | Crystal structure of putative metal-dependent hydrolase APC36150 | | Descriptor: | NICKEL (II) ION, Putative metal-dependent hydrolase | | Authors: | Cooper, D.R, Grelewska, K, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-03-19 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | The structure of DinB from Geobacillus stearothermophilus: a representative of a unique four-helix-bundle superfamily.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3U3K
 
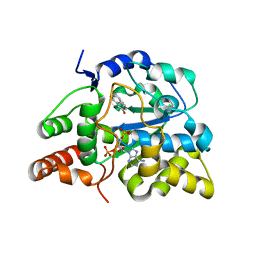 | | Crystal structure of hSULT1A1 bound to PAP and 2-Naphtol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1, naphthalen-2-ol | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3GWW
 
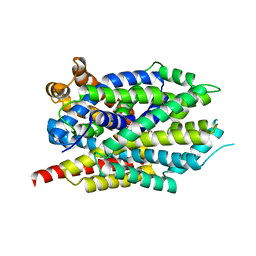 | | Leucine transporter LeuT in complex with S-fluoxetine | | Descriptor: | (3S)-N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, LEUCINE, SODIUM ION, ... | | Authors: | Zhou, Z, Zhen, J, Karpowich, N.K, Law, C.J, Reith, M.E.A, Wang, D.N. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4UZ9
 
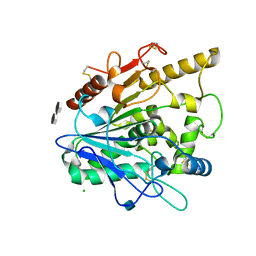 | | STRUCTURE OF THE WNT DEACYLASE NOTUM - CRYSTAL FORM VII - SOS COMPLEX - 2.2A | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Notum Deacylates Wnt Proteins to Suppress Signalling Activity.
Nature, 519, 2015
|
|
3UAZ
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with inosine | | Descriptor: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3GYP
 
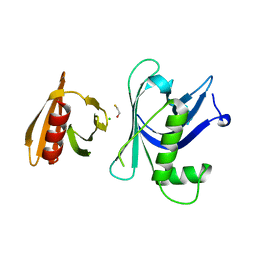 | | Rtt106p | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Histone chaperone RTT106 | | Authors: | Liu, Y, Huang, H, Shi, Y, Teng, M. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
3UBC
 
 | | Oxygen-bound hell's gate globin I by LB nanotemplate method | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Belmonte, L, Scudieri, D, Nicolini, C, Pechkova, E. | | Deposit date: | 2011-10-24 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Oxygen-bound Hell's gate globin I by classical versus LB nanotemplate method.
J.Cell.Biochem., 8, 2012
|
|
6ZR0
 
 | |
3GZ2
 
 | | Crystal structure of IpgC in complex with an IpaB peptide | | Descriptor: | Chaperone protein ipgC, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lokareddy, R.K, Lunelli, M, Kolbe, M. | | Deposit date: | 2009-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combination of two separate binding domains defines stoichiometry between type III secretion system chaperone IpgC and translocator protein IpaB
J.Biol.Chem., 285, 2010
|
|
6ZR4
 
 | |
3U6J
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyrazolone inhibitor | | Descriptor: | N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
7LLK
 
 | |
3GZG
 
 | |
