7XRL
 
 | | Diol dehydratase complexed with AdoMeCbl and 1,2-propanediol | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Shibata, N, Toraya, T. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-29 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into the Very Low Activity of the Homocoenzyme B 12 Adenosylmethylcobalamin in Coenzyme B 12 -Dependent Diol Dehydratase and Ethanolamine Ammonia-Lyase.
Chemistry, 28, 2022
|
|
7XUC
 
 | | Structure of G9a in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-phenylazanyl-hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
8OHO
 
 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry H11 | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, Cyclic di-AMP synthase CdaA, MAGNESIUM ION, ... | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8OJ6
 
 | | HSV-1 DNA polymerase-processivity factor complex in pre-translocation state | | Descriptor: | DNA (22-MER), DNA (48-MER), DNA polymerase catalytic subunit, ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
9CHO
 
 | | Autoinhibited full-length LRRK2(I2020T) on microtubules with MLi-2 | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Chen, S, Villa, E, Leschziner, A.E. | | Deposit date: | 2024-07-01 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron tomography reveals the microtubule-bound form of inactive LRRK2.
Biorxiv, 2024
|
|
9FDC
 
 | | Co-crystal structure of Galectin-3 with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-~{N}-[3,5-bis(chloranyl)phenyl]-6-(hydroxymethyl)-~{N}-methyl-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-2-carboxamide, Galectin-3, NONAETHYLENE GLYCOL, ... | | Authors: | Mac Sweeney, A. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-26 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of Galactopyranose-1-carboxamides as a New Class of Small, Novel, Potent, Selective, and Orally Active Galectin-3 Inhibitors.
Chemmedchem, 20, 2025
|
|
8OJA
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
9FDB
 
 | | Co-crystal structure of Galectin-3 with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-~{N}-cyclopropyl-~{N}-(2-hydroxyethyl)-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-2-carboxamide, Galectin-3, THIOCYANATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2024-05-16 | | Release date: | 2025-03-26 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Galactopyranose-1-carboxamides as a New Class of Small, Novel, Potent, Selective, and Orally Active Galectin-3 Inhibitors.
Chemmedchem, 20, 2025
|
|
6HDH
 
 | |
8AHW
 
 | | Structure of DCS-resistant variant D322N of alanine racemase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Alanine racemase, GLYCEROL | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8REL
 
 | |
8HS5
 
 | |
8OJD
 
 | | HSV-1 DNA polymerase beta-hairpin loop | | Descriptor: | CALCIUM ION, DNA (47-MER), DNA (68-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
8R7B
 
 | | SARS-CoV-2 NSP14 in complex with SAH and TDI-015051 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Meyer, C, Garzia, A, Miller, M, Huggins, D.J, Myers, R.W, Liverton, N, Kargman, S, Nitsche, J, Ganichkin, O, Steinbacher, S, Meinke, P.T, Tuschl, T. | | Deposit date: | 2023-11-24 | | Release date: | 2024-10-23 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Small-molecule inhibition of SARS-CoV-2 NSP14 RNA cap methyltransferase.
Nature, 637, 2025
|
|
6EIJ
 
 | | DYRK1A in complex with HG-8-60-1 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}-[5-[3-(2-morpholin-4-ylethylcarbamoylamino)phenyl]-[1,3]thiazolo[5,4-b]pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Rothweiler, U. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Novel Scaffolds for Dual Specificity Tyrosine-Phosphorylation-Regulated Kinase (DYRK1A) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EMN
 
 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2017-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
5MAJ
 
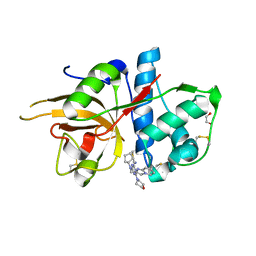 | | CATHEPSIN L IN COMPLEX WITH 4-[cyclopentyl(imidazo[1,2-a]pyridin-2-ylmethyl)amino]-6-morpholino-1,3,5-triazine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, ~{N}-cyclopentyl-~{N}-(imidazo[1,2-a]pyridin-2-ylmethyl)-4-(iminomethyl)-6-morpholin-4-yl-1,3,5-triazin-2-amine | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
6Q7H
 
 | |
7NWL
 
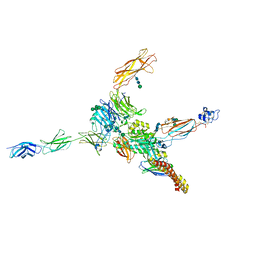 | | Cryo-EM structure of human integrin alpha5beta1 (open form) in complex with fibronectin and TS2/16 Fv-clasp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-5, ... | | Authors: | Schumacher, S, Dedden, D, Vazquez Nunez, R, Matoba, K, Takagi, J, Biertumpfel, C, Mizuno, N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into integrin alpha 5 beta 1 opening by fibronectin ligand.
Sci Adv, 7, 2021
|
|
7UOS
 
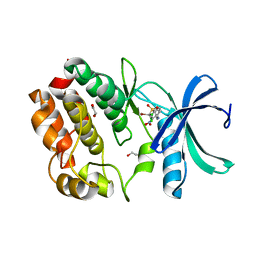 | | Structure of WNK1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 4-[bromo(dichloro)methanesulfonyl]-N-cyclohexyl-2-nitroaniline, Serine/threonine-protein kinase WNK1 | | Authors: | Akella, R, Goldsmith, E.J, Akella, R. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of WNK1 inhibitor complex
To Be Published
|
|
8R71
 
 | | Polysaccharide lyase BtPL33HA (BT4410) Y291A with HAdp4 collected at 1.22 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparinase, ZINC ION, ... | | Authors: | Cartmell, A. | | Deposit date: | 2023-11-23 | | Release date: | 2024-12-04 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Bacterial polysaccharide lyase family 33: Specificity from an evolutionarily conserved binding tunnel.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7F1Y
 
 | | L-lactate oxidase without substrate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Morimoto, Y, Inaka, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Dynamic interactions in the l-lactate oxidase active site facilitate substrate binding at pH4.5.
Biochem.Biophys.Res.Commun., 568, 2021
|
|
7T1O
 
 | |
5FUL
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with SAH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
4ZFK
 
 | | Ergothioneine-biosynthetic Ntn hydrolase EgtC with glutamine | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase EgtC, GLUTAMINE | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the Ergothioneine-Biosynthesis Amidohydrolase EgtC.
Chembiochem, 16, 2015
|
|
