1UBS
 
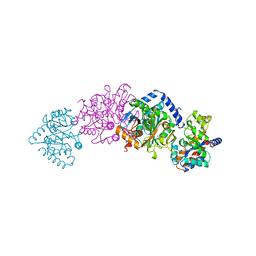 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) WITH A MUTATION OF LYS 87->THR IN THE B SUBUNIT AND IN THE PRESENCE OF LIGAND L-SERINE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SODIUM ION, ... | | Authors: | Rhee, S, Parris, K, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
1UBT
 
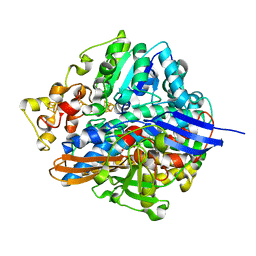 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UBU
 
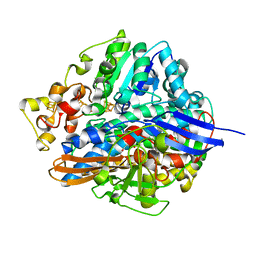 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UBV
 
 | |
1UBW
 
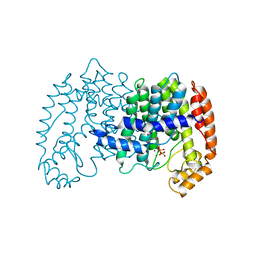 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE SYNTHASE, GERANYL DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBX
 
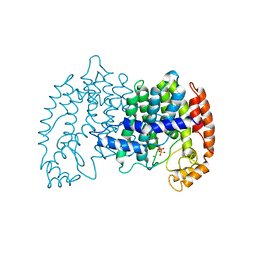 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | FARNESYL DIPHOSPHATE, FARNESYL DIPHOSPHATE SYNTHASE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBY
 
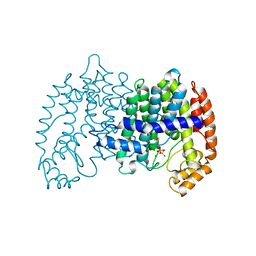 | | STRUCTURE OF FARNESYL PYROPHOSPHATE SYNTHETASE | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, FARNESYL DIPHOSPHATE SYNTHASE, MAGNESIUM ION | | Authors: | Tarshis, L.C, Proteau, P, Poulter, C.D, Sacchettini, J.C. | | Deposit date: | 1996-10-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of product chain length by isoprenyl diphosphate synthases.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1UBZ
 
 | |
1UC0
 
 | |
1UC2
 
 | | Hypothetical Extein Protein of PH1602 from Pyrococcus horikoshii | | Descriptor: | SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, hypothetical protein PH1602 | | Authors: | Okada, C, Maegawa, Y, Yao, M, Tanaka, I. | | Deposit date: | 2003-04-08 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold
Proteins, 63, 2006
|
|
1UC3
 
 | |
1UC4
 
 | | Structure of diol dehydratase complexed with (S)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.BIOL.CHEM., 278, 2003
|
|
1UC5
 
 | | Structure of diol dehydratase complexed with (R)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.Biol.Chem., 278, 2003
|
|
1UC6
 
 | | Solution Structure of the Carboxyl Terminal Domain of the Ciliary Neurotrophic Factor Receptor | | Descriptor: | Ciliary Neurotrophic Factor Receptor alpha | | Authors: | Man, D, He, W, Sze, K.H, Ke, G, Smith, D.K, Ip, N.Y, Zhu, G. | | Deposit date: | 2003-04-08 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of the ciliary neurotrophic factor (CNTF) receptor and ligand free associations among components of the CNTF receptor complex
J.Biol.Chem., 278, 2003
|
|
1UC7
 
 | | Crystal structure of DsbDgamma | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Kim, J.H, Kim, S.J, Jeong, D.G, Son, J.H, Ryu, S.E. | | Deposit date: | 2003-04-09 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of DsbDgamma reveals the mechanism of redox potential shift and substrate specificity(1)
FEBS LETT., 543, 2003
|
|
1UC8
 
 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
1UC9
 
 | | Crystal structure of a lysine biosynthesis enzyme, Lysx, from thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, lysine biosynthesis enzyme | | Authors: | Sakai, H, Vassylyeva, M.N, Matsuura, T, Sekine, S, Nishiyama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Lysine Biosynthesis Enzyme, LysX, from Thermus thermophilus HB8
J.Mol.Biol., 332, 2003
|
|
1UCA
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 2'-UMP | | Descriptor: | PHOSPHORIC ACID MONO-[2-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-4-HYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1UCB
 
 | |
1UCC
 
 | | Crystal structure of the Ribonuclease MC1 from bitter gourd seeds complexed with 3'-UMP. | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease MC | | Authors: | Suzuki, A, Yao, M, Tanaka, I, Numata, T, Kikukawa, S, Yamasaki, N, Kimura, M. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2'-UMP or 3'-UMP, reveal structural basis for uridine specificity
Biochem.Biophys.Res.Commun., 275, 2000
|
|
1UCD
 
 | | Crystal structure of Ribonuclease MC1 from bitter gourd seeds complexed with 5'-UMP | | Descriptor: | Ribonuclease MC, URACIL, URIDINE-5'-MONOPHOSPHATE | | Authors: | Suzuki, A, Numata, T, Yao, M, Kimura, M, Tanaka, I. | | Deposit date: | 2003-04-10 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of RNase MC1 from bitter gourd seeds in complex with 5'UMP
To be published
|
|
1UCF
 
 | | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Honbou, K, Suzuki, N.N, Horiuchi, M, Niki, T, Taira, T, Ariga, H, Inagaki, F. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease
J.BIOL.CHEM., 278, 2003
|
|
1UCG
 
 | | Crystal structure of Ribonuclease MC1 N71T mutant | | Descriptor: | MANGANESE (II) ION, Ribonuclease MC | | Authors: | Suzuki, A, Numata, T, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2003-04-14 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity
Biochemistry, 42, 2003
|
|
1UCH
 
 | | DEUBIQUITINATING ENZYME UCH-L3 (HUMAN) AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | UBIQUITIN C-TERMINAL HYDROLASE UCH-L3 | | Authors: | Johnston, S.C, Larsen, C.N, Cook, W.J, Wilkinson, K.D, Hill, C.P. | | Deposit date: | 1997-10-06 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution.
EMBO J., 16, 1997
|
|
1UCI
 
 | | Mutants of RNase Sa | | Descriptor: | Guanyl-specific ribonuclease Sa, SULFATE ION | | Authors: | Takano, K, Scholtz, J.M, Sacchettini, J.C, Pace, C.N. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The contribution of polar group burial to protein stability is strongly context-dependent
J.Biol.Chem., 278, 2003
|
|
