4EV2
 
 | |
1O6U
 
 | |
1O7D
 
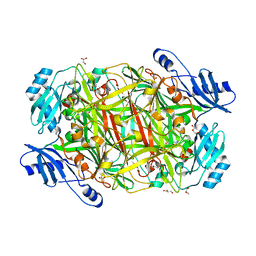
 | | The structure of the bovine lysosomal a-mannosidase suggests a novel mechanism for low pH activation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Heikinheimo, P, Helland, R, Leiros, H.S, Leiros, I, Karlsen, S, Evjen, G, Ravelli, R, Schoehn, G, Ruigrok, R, Tollersrud, O.-K, Mcsweeney, S, Hough, E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of Bovine Lysosomal Alpha-Mannosidase Suggests a Novel Mechanism for Low-Ph Activation
J.Mol.Biol., 327, 2003
|
|
1OBK
 
 | |
1OCM
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COVALENTLY COMPLEXED WITH PYROPHOSPHATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, PYROPHOSPHATE 2- | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OD9
 
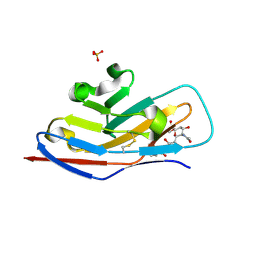 | | N-terminal of Sialoadhesin in complex with Me-a-9-N-benzoyl-amino-9-deoxy-Neu5Ac (BENZ compound) | | Descriptor: | SIALOADHESIN, SULFATE ION, methyl 5-acetamido-3,5,9-trideoxy-9-[(phenylcarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosidonic acid | | Authors: | Zaccai, N.R, Maenaka, K, Maenaka, T, Crocker, P.R, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2003-02-14 | | Release date: | 2003-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of Sialic Acid-Based Siglec Inhibitors and Crystallographic Analysis in Complex with Sialoadhesin
Structure, 11, 2003
|
|
1O6P
 
 | |
1O9N
 
 | |
1ODH
 
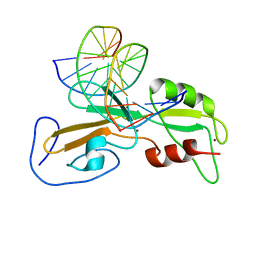 | | Structure of the GCM domain bound to DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*GP*CP*GP*GP*GP*TP *GP*CP*A)-3', 5'-D(*TP*GP*CP*AP*CP*CP*CP*GP*CP*AP *TP*CP*G)-3', MGCM1, ... | | Authors: | Cohen, S.X, Muller, C.W. | | Deposit date: | 2003-02-19 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of the Gcm Domain-DNA Complex: A DNA-Binding Domain with a Novel Fold and Mode of Target Site Recognition
Embo J., 22, 2003
|
|
1OA9
 
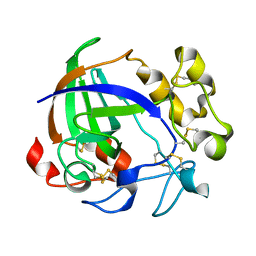 | |
1NSG
 
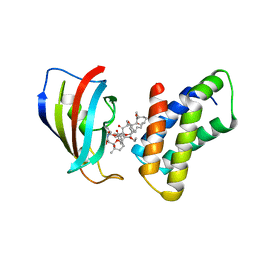 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FKBP-RAPAMYCIN ASSOCIATED PROTEIN (FRAP) | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1NPX
 
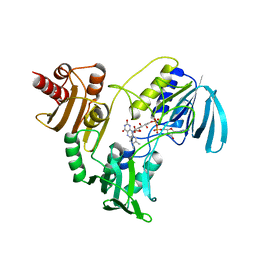 | | STRUCTURE OF NADH PEROXIDASE FROM STREPTOCOCCUS FAECALIS 10C1 REFINED AT 2.16 ANGSTROMS RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE | | Authors: | Stehle, T, Ahmed, S.A, Claiborne, A, Schulz, G.E. | | Deposit date: | 1991-08-02 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of NADH peroxidase from Streptococcus faecalis 10C1 refined at 2.16 A resolution.
J.Mol.Biol., 221, 1991
|
|
1OAP
 
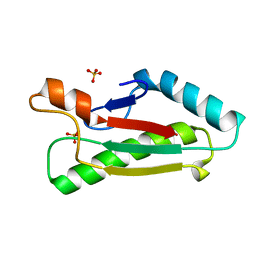 | | Mad structure of the periplasmique domain of the Escherichia coli PAL protein | | Descriptor: | PEPTIDOGLYCAN-ASSOCIATED LIPOPROTEIN, SULFATE ION | | Authors: | Abergel, C, Walburger, A, Bouveret, E, Claverie, J.M. | | Deposit date: | 2003-01-20 | | Release date: | 2004-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallization and preliminary crystallographic study of the peptidoglycan-associated lipoprotein from Escherichia coli.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
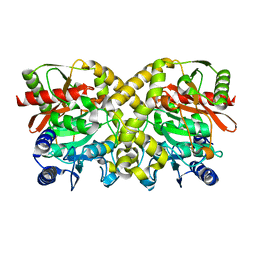
1OBI
 
 | |
1NSK
 
 | |
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
1NP1
 
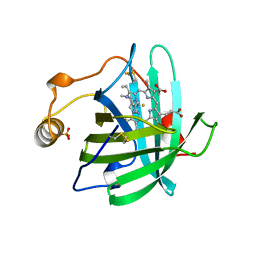 | |
1NR7
 
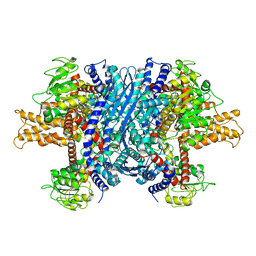 | | Crystal structure of apo bovine glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1O16
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN H64D/V68S/D122N MUTANT (MET) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N. | | Deposit date: | 2002-10-25 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular engineering of myoglobin: influence of residue 68 on the rate and the
enantioselectivity of oxidation reactions catalyzed by H64D/V68X myoglobin
Biochemistry, 42, 2003
|
|
1O4N
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH OXALIC ACID. | | Descriptor: | OXALIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O5W
 
 | | The structure basis of specific recognitions for substrates and inhibitors of rat monoamine oxidase A | | Descriptor: | Amine oxidase [flavin-containing] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | Ma, J, Yoshimura, M, Yamashita, E, Nakagawa, A, Ito, A, Tsukihara, T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of rat monoamine oxidase a and its specific recognitions for substrates and inhibitors.
J.Mol.Biol., 338, 2004
|
|
1PX8
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1PYX
 
 | | GSK-3 Beta complexed with AMP-PNP | | Descriptor: | Glycogen synthase kinase-3 beta, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of the GSK-3beta active site using selective and non-selective ATP-mimetic inhibitors
J.Mol.Biol., 333, 2003
|
|
1Q2Y
 
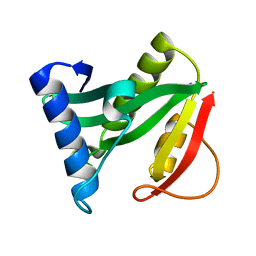 | | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily fold | | Descriptor: | similar to hypothetical proteins | | Authors: | Fedorov, A.A, Ramagopal, U.A, Fedorov, E.V, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-07-27 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily
To be Published
|
|
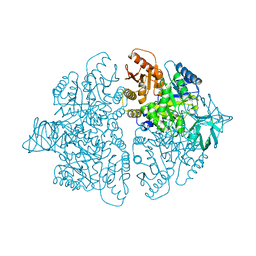
1PKM
 
 | |
