1MPN
 
 | | MALTOPORIN MALTOTRIOSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1996-01-11 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway.
Structure, 4, 1996
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1M00
 
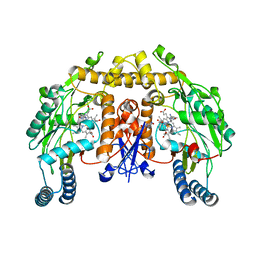 | | Rat neuronal NOS heme domain with N-butyl-N'-hydroxyguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-BUTYL-N'-HYDROXYGUANIDINE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
1M0S
 
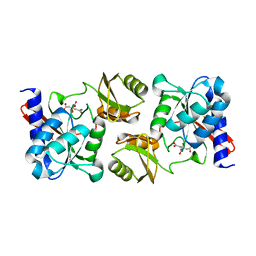 | | NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG ID IR21) | | Descriptor: | CITRIC ACID, Ribose-5-Phosphate Isomerase A | | Authors: | Das, K, Xiao, R, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-14 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-RIBOSE-5-PHOSPHATE ISOMERASE, IR21
TO BE PUBLISHED
|
|
1LYC
 
 | | The impact of the physical and chemical enviroment on the molecular structure of Coprinus cinereus peroxidase | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Houborg, K, Harris, P, Petersen, J.F.W, Rowland, P, Poulsen, J.-C.N, Schneider, P, Vind, J, Larsen, S. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Impact of the physical and chemical environment on the molecular structure of Coprinus cinereus peroxidase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1M2I
 
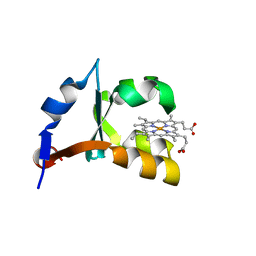 | | Crystal structure of E44A/E56A mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, J, Wang, Y.-H, Gan, J.-H, Wang, W.-H, Sun, B.-Y, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-06-24 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Cytochrome b5 Mutated at the Charged Surface-Residues and Their Interactions with Cytochrome c
Chin.J.Chem., 20, 2002
|
|
1M2Q
 
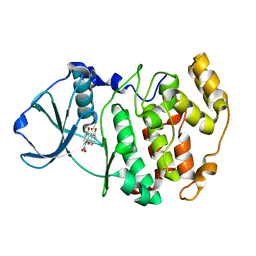 | | Crystal structure of 1,8-di-hydroxy-4-nitro-xanten-9-one/CK2 kinase complex | | Descriptor: | 1,8-DI-HYDROXY-4-NITRO-XANTHEN-9-ONE, Casein kinase II, alpha chain | | Authors: | De Moliner, E, Sarno, S, Moro, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
1M3W
 
 | | Crystal Structure of a Molecular Maquette Scaffold | | Descriptor: | H10H24, MERCURY (II) ION | | Authors: | Huang, S.S, Gibney, B.R, Stayrook, S.E, Dutton, P.L, Lewis, M. | | Deposit date: | 2002-07-01 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Structure of a Maquette Scaffold
J.Mol.Biol., 326, 2003
|
|
1PTF
 
 | |
1PM3
 
 | |
1PYG
 
 | |
1PPE
 
 | |
1PXT
 
 | |
1PU9
 
 | | Crystal Structure of Tetrahymena GCN5 with Bound Coenzyme A and a 19-residue Histone H3 Peptide | | Descriptor: | COENZYME A, HAT A1, Histone H3 | | Authors: | Clements, A, Poux, A.N, Lo, W.S, Pillus, L, Berger, S.L, Marmorstein, R. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for histone and phospho-histone binding by the GCN5 histone acetyltransferase
Mol.Cell, 12, 2003
|
|
1EB1
 
 | | Complex structure of human thrombin with N-methyl-arginine inhibitor | | Descriptor: | 3-CYCLOHEXYL-D-ALANYL-L-PROLYL-N~2~-METHYL-L-ARGININE, PEPTIDE INHIBITOR, THROMBIN HEAVY CHAIN, ... | | Authors: | Friedrich, R, Steinmetzer, T, Bode, W. | | Deposit date: | 2001-07-18 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Methyl Group of N(Alpha)(Me)Arg-Containing Peptides Disturbs the Active-Site Geometry of Thrombin, Impairing Efficient Cleavage
J.Mol.Biol., 316, 2002
|
|
1PVT
 
 | | Crystal structure of sugar-phosphate aldolase from Thermotoga maritima | | Descriptor: | sugar-phosphate aldolase | | Authors: | Osipiuk, J, Cuff, M.E, Korolev, O, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-28 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of sugar-phosphate aldolase from Thermotoga maritima.
To be Published
|
|
1PLF
 
 | |
1Q09
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1PNZ
 
 | |
1Q17
 
 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, HST2 protein, ... | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Yeast Hst2 Protein Deacetylase in Ternary Complex with 2'-O-Acetyl
ADP Ribose and Histone Peptide.
Structure, 11, 2003
|
|
1PT3
 
 | | Crystal structures of nuclease-ColE7 complexed with octamer DNA | | Descriptor: | 5'-GCGATCGC-3', Colicin E7 | | Authors: | Hsia, K.C, Chak, K.F, Cheng, Y.S, Ku, W.Y, Yuan, H.S. | | Deposit date: | 2003-06-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA binding and degradation by the HNH protein ColE7.
STRUCTURE, 12, 2004
|
|
1Q3R
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1PTS
 
 | |
1PWO
 
 | |
1PPN
 
 | | STRUCTURE OF MONOCLINIC PAPAIN AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, PAPAIN, UNKNOWN LIGAND | | Authors: | Pickersgill, R.W, Harris, G.W, Garman, E. | | Deposit date: | 1991-10-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Monoclinic Papain at 1.60 Angstroms Resolution
Acta Crystallogr.,Sect.B, 48, 1992
|
|
