7YOE
 
 | |
8PVS
 
 | | Targeting extended blood antigens by Akkermansia muciniphila enzymes unveils a missing link for generating universal donor blood | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3-galactosidase B, CALCIUM ION, ... | | Authors: | Jensen, M, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2023-07-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Akkermansia muciniphila exoglycosidases target extended blood group antigens to generate ABO-universal blood.
Nat Microbiol, 9, 2024
|
|
5NTU
 
 | | Crystal Structure of human Pro-myostatin Precursor at 2.6 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Growth/differentiation factor 8 | | Authors: | Cotton, T.R, Fischer, G, Hyvonen, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human myostatin precursor and determinants of growth factor latency.
EMBO J., 37, 2018
|
|
7SV1
 
 | |
7SV8
 
 | |
5HVM
 
 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase A in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.815 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
7YOD
 
 | |
7SUW
 
 | |
6FCF
 
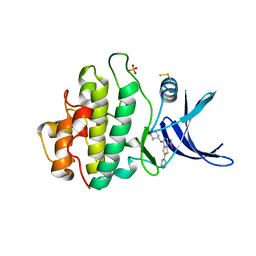 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 44 | | Descriptor: | 4-[[(2~{R},3~{S})-2-methylpiperidin-3-yl]amino]-2-phenyl-thieno[3,2-c]pyridine-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
8BTO
 
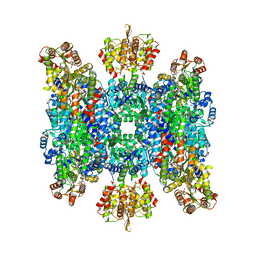 | | Helical structure of BcThsA in complex with 1''-3'gcADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, NAD(+) hydrolase ThsA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
7JJT
 
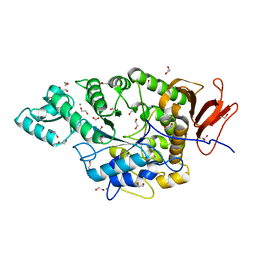 | | Ruminococcus bromii amylase Amy5 (RBR_07800) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha-amylase, ... | | Authors: | Cerqueira, F, Koropatkin, N. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The structures of the GH13_36 amylases from Eubacterium rectale and Ruminococcus bromii reveal subsite architectures that favor maltose production
Amylase, 4, 2020
|
|
5NWA
 
 | |
5JJR
 
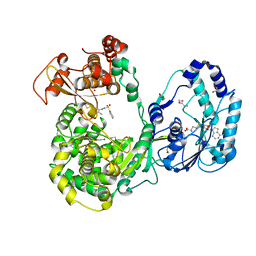 | | Dengue 3 NS5 protein with compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-[5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl]-4-methoxy-2-methyl-N-[(quinolin-8-yl)sulfonyl]benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lescar, J, El Sahili, A. | | Deposit date: | 2016-04-25 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling
Plos Pathog., 12, 2016
|
|
6F8X
 
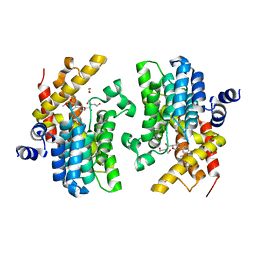 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-26g | | Descriptor: | 1,2-ETHANEDIOL, 2-[(5~{R})-3-(3-cyclopentyloxy-4-methoxy-phenyl)-4,5-dihydro-1,2-oxazol-5-yl]-~{N},~{N}-bis(2-hydroxyethyl)ethanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F28
 
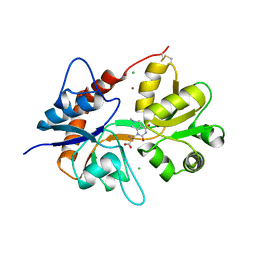 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2'-Amino-2'-carboxyethyl]-6-methyl-5,7-dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.4A | | Descriptor: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-11-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
8BT7
 
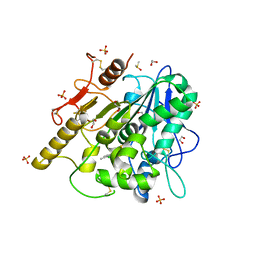 | | Notum Inhibitor ARUK3004903 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3,4-bis(chloranyl)-5-methyl-indol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
4YOK
 
 | |
6BVY
 
 | | SFTI-HFRW-4 | | Descriptor: | Trypsin inhibitor 1 HFRW-4 | | Authors: | Schroeder, C.I, White, A. | | Deposit date: | 2017-12-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J. Med. Chem., 61, 2018
|
|
5HXA
 
 | |
8BTC
 
 | | Notum Inhibitor ARUK3004558 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4,5-bis(chloranyl)-2,3-dihydroindol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8T1Q
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8BTI
 
 | | Notum Inhibitor ARUK3004556 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-chloranylindol-1-yl)-2-methoxy-ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT2
 
 | | Notum Inhibitor ARUK3004876 | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-chloranyl-4-(trifluoromethyl)-2,3-dihydroindol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTA
 
 | | Notum Inhibitor ARUK3004308 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-chloranyl-2,3-dihydroindol-1-yl)ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
1EEZ
 
 | |
