2YL7
 
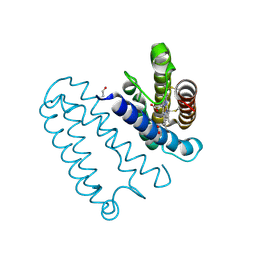 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: AS ISOLATED L16G VARIANT AT 0.9 A RESOLUTION - RESTRAINT REFINEMENT | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2YLG
 
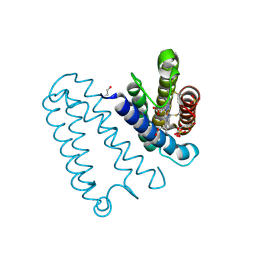 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: ASCORBATE AND CARBON MONOOXIDE BOUND L16A VARIANT AT 1.05 A RESOLUTION | | Descriptor: | ASCORBIC ACID, CARBON MONOXIDE, CYTOCHROME C', ... | | Authors: | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2011-06-02 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4E4W
 
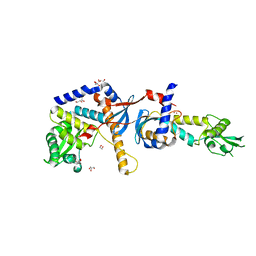 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-03-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1UOB
 
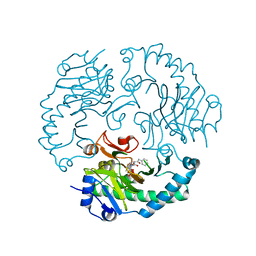 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and penicillin G | | Descriptor: | 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UNB
 
 | | Deacetoxycephalosporin C synthase complexed with 2-oxoglutarate and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, ... | | Authors: | Valegard, K, Terwisscha van Scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-09 | | Release date: | 2004-01-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
3ZOO
 
 | |
2YLI
 
 | |
4GED
 
 | | Crystal Structure of the Leishmania Major Peroxidase-Cytochrome C Complex | | Descriptor: | Ascorbate peroxidase, Cytochrome c, HEME C, ... | | Authors: | Jasion, V.S, Doukov, T, Pineda, S.H, Li, H, Poulos, T.L. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the Leishmania major peroxidase-cytochrome c complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5CIB
 
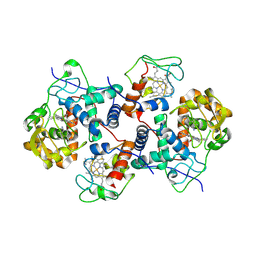 | | Complex of yeast cytochrome c peroxidase (W191G) bound to 2,4-dimethylaniline with iso-1 cytochrome c | | Descriptor: | 2,4-dimethylaniline, Cytochrome c iso-1, Cytochrome c peroxidase, ... | | Authors: | Crane, B.R, Payne, T.M. | | Deposit date: | 2015-07-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Constraints on the Radical Cation Center of Cytochrome c Peroxidase for Electron Transfer from Cytochrome c.
Biochemistry, 55, 2016
|
|
7EI4
 
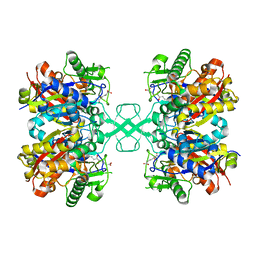 | | Crystal structure of MasL in complex with a novel covalent inhibitor, collimonin C | | Descriptor: | (6S,7R,9E)-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
3U6H
 
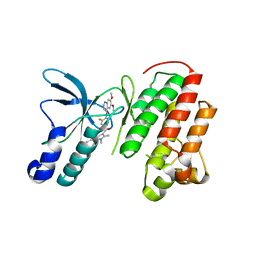 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 26 | | Descriptor: | Hepatocyte growth factor receptor, N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3OB2
 
 | |
1YEB
 
 | |
3WHD
 
 | | C-type lectin, human MCL | | Descriptor: | C-type lectin domain family 4 member D, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GYX
 
 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1SV1
 
 | |
4GRU
 
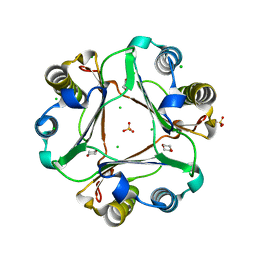 | |
2CEP
 
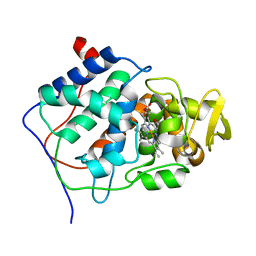 | |
1E8E
 
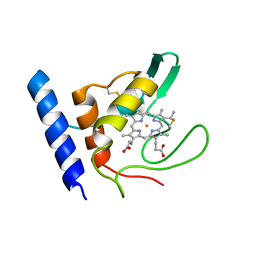 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
2GO4
 
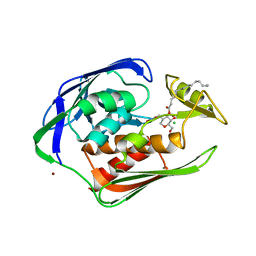 | | Crystal structure of Aquifex aeolicus LpxC complexed with TU-514 | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
4GRQ
 
 | | Characterization of N- and C- terminus mutants of human MIF | | Descriptor: | 2-methyl-1-[2-(propan-2-yl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor, ... | | Authors: | Fan, C, Lolis, E. | | Deposit date: | 2012-08-26 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of N- and C- terminus mutants of human MIF
To be Published
|
|
4GRO
 
 | |
2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
3CU8
 
 | | Impaired binding of 14-3-3 to Raf1 is linked to Noonan and LEOPARD syndrome | | Descriptor: | 14-3-3 protein zeta/delta, MAGNESIUM ION, PROPANOIC ACID, ... | | Authors: | Schumacher, B, Weyand, M, Kuhlmann, J, Ottmann, C. | | Deposit date: | 2008-04-16 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol. Cell. Biol., 30, 2010
|
|
