1TZN
 
 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore bound to the VWA domain of CMG2, an anthrax toxin receptor | | Descriptor: | Anthrax toxin receptor 2, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | Deposit date: | 2004-07-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TZO
 
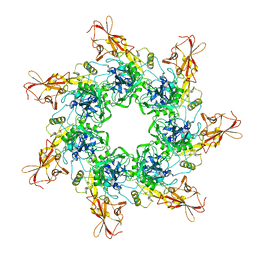 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | Deposit date: | 2004-07-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TZP
 
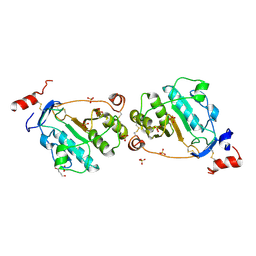 | | MEPA, inactive form without ZN in P21 | | Descriptor: | 1,4-BUTANEDIOL, Penicillin-insensitive murein endopeptidase, SULFATE ION | | Authors: | Marcyjaniak, M, Odintsov, S.G, Sabala, I, Bochtler, M. | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptidoglycan amidase MepA is a LAS metallopeptidase
J.Biol.Chem., 279, 2004
|
|
1TZQ
 
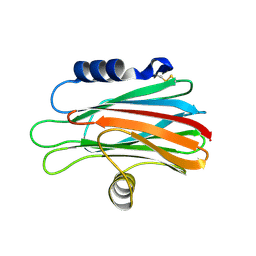 | | Crystal structure of the equinatoxin II 8-69 double cysteine mutant | | Descriptor: | Equinatoxin II | | Authors: | Kristan, K, Podlesek, Z, Hojnik, V, Gutirrez-Aguirre, I, Guncar, G, Turk, D.A, Gonzalez-Maas, J.M, Lakey, J.H, Anderluh, G. | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pore formation by equinatoxin, a eukaryotic pore-forming toxin, requires a flexible N-terminal region and a stable beta-sandwich
J.Biol.Chem., 279, 2004
|
|
1TZS
 
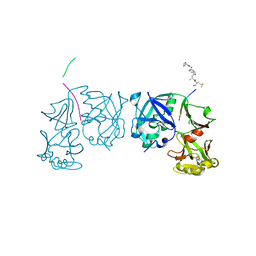 | | Crystal Structure of an activation intermediate of Cathepsin E | | Descriptor: | 23-mer peptide from PelB-IgG kappa light chain fusion protein, Cathepsin E, activation peptide from Cathepsin E | | Authors: | Ostermann, N, Gerhartz, B, Worpenberg, S, Trappe, J, Eder, J. | | Deposit date: | 2004-07-12 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of an activation intermediate of cathepsin e
J.Mol.Biol., 342, 2004
|
|
1TZT
 
 | | T. maritima NusB, P21 | | Descriptor: | N utilization substance protein B homolog, SULFATE ION | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
1TZU
 
 | | T. maritima NusB, P212121 | | Descriptor: | N utilization substance protein B homolog, SULFATE ION | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
1TZV
 
 | | T. maritima NusB, P3121, Form 1 | | Descriptor: | N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
1TZW
 
 | | T. maritima NusB, P3121, Form 2 | | Descriptor: | CALCIUM ION, N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
1TZX
 
 | | T. maritima NusB, P3221 | | Descriptor: | CITRIC ACID, N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
1TZY
 
 | | Crystal Structure of the Core-Histone Octamer to 1.90 Angstrom Resolution | | Descriptor: | CHLORIDE ION, HISTONE H3, HISTONE H4-VI, ... | | Authors: | Wood, C.M, Nicholson, J.M, Chantalat, L, Reynolds, C.D, Lambert, S.J, Baldwin, J.P. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of the native histone octamer.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1TZZ
 
 | | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum | | Descriptor: | Hypothetical protein L1841, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum
To be Published
|
|
1U00
 
 | | HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC | | Descriptor: | Chaperone protein hscA, IscU recognition peptide | | Authors: | Cupp-Vickery, J.R, Peterson, J.C, Ta, D.T, Vickery, L.E. | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Molecular Chaperone HscA Substrate Binding Domain Complexed with the IscU Recognition Peptide ELPPVKIHC.
J.Mol.Biol., 342, 2004
|
|
1U01
 
 | | High resolution NMR structure of 5-d(GCGT*GCG)-3/5-d(CGCACGC)-3 (T*represents a cyclohexenyl nucleotide) | | Descriptor: | 5'-D(*CP*GP*CP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*(XTR)P*GP*CP*G)-3' | | Authors: | Nauwelaerts, K, Lescrinier, E, Sclep, G, Herdewijn, P. | | Deposit date: | 2004-07-12 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Cyclohexenyl nucleic acids: conformationally flexible oligonucleotides.
Nucleic Acids Res., 33, 2005
|
|
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
1U04
 
 | |
1U05
 
 | |
1U06
 
 | | crystal structure of chicken alpha-spectrin SH3 domain | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Chevelkov, V, Faelber, K, Diehl, A, Heinemann, U, Oschkinat, H, Reif, B. | | Deposit date: | 2004-07-13 | | Release date: | 2005-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detection of dynamic water molecules in a microcrystalline sample of the SH3 domain of alpha-spectrin by MAS solid-state NMR.
J.Biomol.Nmr, 31, 2005
|
|
1U07
 
 | | Crystal Structure of the 92-residue C-term. part of TonB with significant structural changes compared to shorter fragments | | Descriptor: | TonB protein | | Authors: | Koedding, J, Killig, F, Polzer, P, Howard, S.P, Diederichs, K, Welte, W. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of a 92-residue c-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments
J.Biol.Chem., 280, 2005
|
|
1U08
 
 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
1U09
 
 | | Footand Mouth Disease Virus RNA-dependent RNA polymerase | | Descriptor: | polyprotein | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2004-07-13 | | Release date: | 2004-08-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of Foot-and-Mouth Disease Virus RNA-dependent RNA Polymerase and Its Complex with a Template-Primer RNA
J.Biol.Chem., 279, 2004
|
|
1U0A
 
 | | Crystal structure of the engineered beta-1,3-1,4-endoglucanase H(A16-M) in complex with beta-glucan tetrasaccharide | | Descriptor: | Beta-glucanase, CALCIUM ION, ZINC ION, ... | | Authors: | Gaiser, O.J, Piotukh, K, Ponnuswamy, M.N, Planas, A, Borriss, R, Heinemann, U. | | Deposit date: | 2004-07-13 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis for the Substrate Specificity of a Bacillus 1,3-1,4-beta-Glucanase
J.Mol.Biol., 357, 2006
|
|
1U0B
 
 | | Crystal structure of cysteinyl-tRNA synthetase binary complex with tRNACys | | Descriptor: | Cysteine--tRNA ligase, ZINC ION, cysteinyl tRNA | | Authors: | Hauenstein, S, Zhang, C.M, Hou, Y.M, Perona, J.J. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shape-selective RNA recognition by cysteinyl-tRNA synthetase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1U0C
 
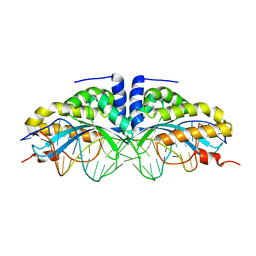 | | Y33C Mutant of Homing endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*TP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*AP*GP*C)-3', 5'-D(*GP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*AP*CP*G)-3', DNA endonuclease I-CreI, ... | | Authors: | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
1U0D
 
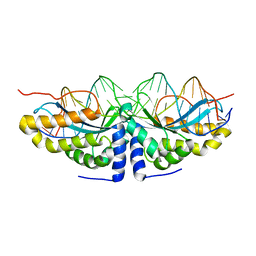 | | Y33H Mutant of Homing endonuclease I-CreI | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', DNA endonuclease I-CreI | | Authors: | Sussman, D, Chadsey, M, Fauce, S, Engel, A, Bruett, A, Monnat, R, Stoddard, B.L, Seligman, L.M. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Isolation and characterization of new homing endonuclease specificities at individual target site positions.
J.Mol.Biol., 342, 2004
|
|
