1OGA
 
 | | A structural basis for immunodominant human T-cell receptor recognition. | | Descriptor: | BETA-2-MICROGLOBULIN, GILGFVFTL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B.E, McMichael, A.J, Bell, J.I, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Structural Basis for Immunodominant Human T Cell Receptor Recognition
Nat.Immunol., 4, 2003
|
|
1OH9
 
 | | Acetylglutamate kinase from Escherichia coli complexed with MgADP, N-acetyl-L-glutamate and the transition-state mimic AlF4- | | Descriptor: | ACETYLGLUTAMATE KINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gil-Ortiz, F, Ramon-Maiques, S, Fita, I, Rubio, V. | | Deposit date: | 2003-05-23 | | Release date: | 2003-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The Course of Phosphorus in the Reaction of N-Acetyl-L-Glutamate Kinase, Determined from the Structures of Crystalline Complexes, Including a Complex with an Alf(4)(-) Transition State Mimic
J.Mol.Biol., 331, 2003
|
|
1P0F
 
 | | Crystal Structure of the Binary Complex: NADP(H)-Dependent Vertebrate Alcohol Dehydrogenase (ADH8) with the cofactor NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent ALCOHOL DEHYDROGENASE, ... | | Authors: | Rosell, A, Valencia, E, Pares, X, Fita, I, Farres, J, Ochoa, W.F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the vertebrate NADP(H)-dependent alcohol dehydrogenase (ADH8)
J.Mol.Biol., 330, 2003
|
|
1P1G
 
 | |
1ORD
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | Deposit date: | 1995-02-08 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
1ONC
 
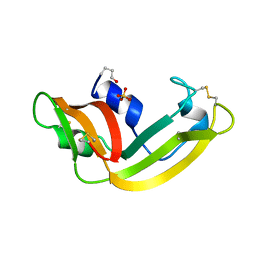 | | THE REFINED 1.7 ANGSTROMS X-RAY CRYSTALLOGRAPHIC STRUCTURE OF P-30, AN AMPHIBIAN RIBONUCLEASE WITH ANTI-TUMOR ACTIVITY | | Descriptor: | P-30 PROTEIN, SULFATE ION | | Authors: | Mosimann, S.C, Ardelt, W, James, M.N.G. | | Deposit date: | 1993-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined 1.7 A X-ray crystallographic structure of P-30 protein, an amphibian ribonuclease with anti-tumor activity.
J.Mol.Biol., 236, 1994
|
|
1OO5
 
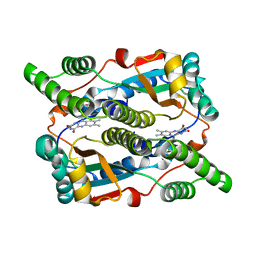 | | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of the Enzyme Active Form and Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-03 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OQE
 
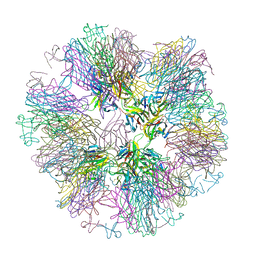 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1P1U
 
 | |
1OSP
 
 | |
1OWF
 
 | | Crystal structure of a mutant IHF (BetaE44A) complexed with the native H' Site | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', Integration Host Factor Alpha-subunit, ... | | Authors: | Lynch, T.W, Read, E.K, Mattis, A.N, Gardner, J.F, Rice, P.A. | | Deposit date: | 2003-03-28 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Integration Host Factor: putting a twist on protein-DNA recognition
J.Mol.Biol., 330, 2003
|
|
1P5Y
 
 | |
1OYO
 
 | | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution | | Descriptor: | 3H-INDOLE-5,6-DIOL, CALCIUM ION, Proteinase K | | Authors: | Singh, N, Sharma, S, Kumar, S, Raman, G, Singh, T.P. | | Deposit date: | 2003-04-06 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution
To be Published
|
|
1P6K
 
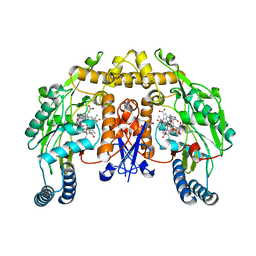 | | Rat neuronal NOS D597N mutant heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P92
 
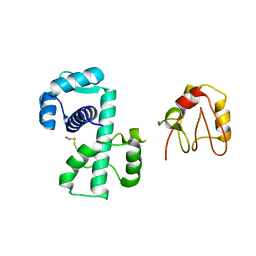 | | Crystal Structure of (H79A)DtxR | | Descriptor: | BETA-MERCAPTOETHANOL, Diphtheria toxin repressor | | Authors: | D'Aquino, J.A, Ringe, D. | | Deposit date: | 2003-05-08 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determinants of the SRC homology domain 3-like fold.
J.Bacteriol., 185, 2003
|
|
1OM1
 
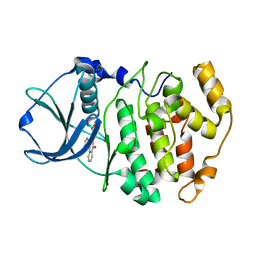 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
1OJO
 
 | |
1OOY
 
 | | SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase, ... | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OPR
 
 | |
1OSU
 
 | |
1ORU
 
 | | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, SULFATE ION, yuaD protein | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-15 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis
To be Published
|
|
1OSV
 
 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1D0K
 
 | | THE ESCHERICHIA COLI LYTIC TRANSGLYCOSYLASE SLT35 IN COMPLEX WITH TWO MURODIPEPTIDES (GLCNAC-MURNAC-L-ALA-D-GLU) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE, ALANINE, ... | | Authors: | van Asselt, E.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-12 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan.
Biochemistry, 39, 2000
|
|
1OSA
 
 | |
1CG0
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH HADACIDIN, GDP, 6-PHOSPHORYL-IMP, AND MG2+ | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Choe, J.Y, Poland, B.W, Fromm, H, Honzatko, R. | | Deposit date: | 1999-03-26 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic implications from crystalline complexes of wild-type and mutant adenylosuccinate synthetases from Escherichia coli.
Biochemistry, 38, 1999
|
|
