7P0K
 
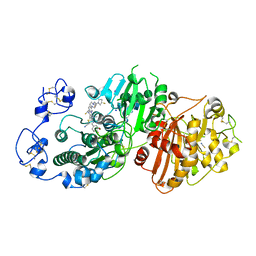 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
1EFV
 
 | |
8P5A
 
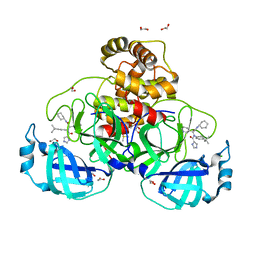 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer R. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
7MX1
 
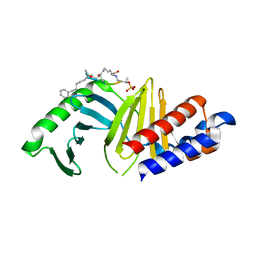 | | PLK-1 polo-box domain in complex with a high affinity macrocycle synthesized using a novel glutamic acid analog | | Descriptor: | ACE-PRO-LEU-ALA-SER-TPO, N-[(4S)-4,5-diamino-5-oxopentyl]-10-phenyldecanamide, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Hymel, D, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2021-05-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of a new orthogonally protected glutamic acid analog and its use in the preparation of high affinity polo-like kinase 1 polo-box domain - binding peptide macrocycles.
Org.Biomol.Chem., 19, 2021
|
|
8P87
 
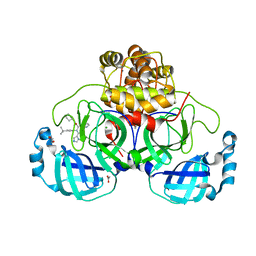 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM X77, from an "old" crystal. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
5GYI
 
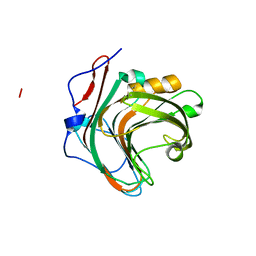 | |
8P56
 
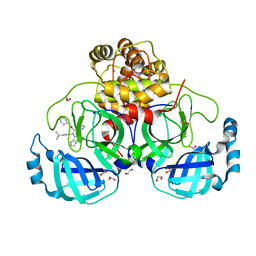 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar X77. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
5GY9
 
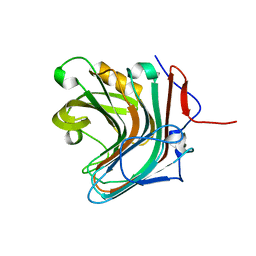 | |
5GYF
 
 | |
8FZ2
 
 | | Crystal structure of Fab460 in complex with MPER peptide | | Descriptor: | Fab460, H chain, L chain, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
8UFK
 
 | |
8UFF
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGGAAGTGGG) in Ternary Complex with DB1976 | | Descriptor: | (2M,2'M)-2,2'-(selenophene-2,5-diyl)di(1H-benzimidazole-6-carboximidamide), DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*TP*TP*TP*TP*AP*T)-3'), ... | | Authors: | Terrell, J.R, Poon, G.M.K, Wilson, W.D. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural studies on the PU.1 inhibitory mechanism by diamidine minor groove binders
To Be Published
|
|
6U7W
 
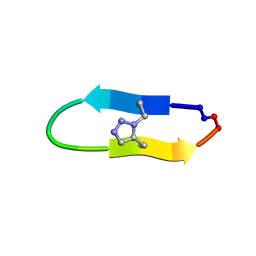 | | NMR solution structure of a triazole bridged KLK7 inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-LYS-ALA-LEU-PHE-SER-ASN-PRO-PRO-ILE-ALA-PHE-PRO-ASN | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6F8J
 
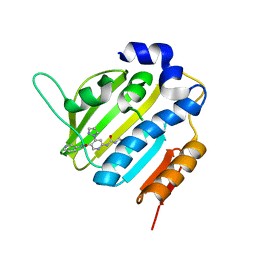 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
8AV1
 
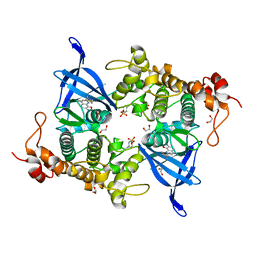 | | Crystal structure of GSK3 beta (GSK3b) in complex with CD7. | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-8-thiomorpholin-4-yl-[1,3]oxazolo[5,4-f]quinoxaline, Glycogen synthase kinase-3 beta, ... | | Authors: | Chaikuad, A, Mongin, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxazolo[5,4-f]quinoxaline-type selective inhibitors of glycogen synthase kinase-3 alpha (GSK-3 alpha ): Development and impact on temozolomide treatment of glioblastoma cells.
Bioorg.Chem., 134, 2023
|
|
8B43
 
 | | Crystal structure of ferrioxamine transporter | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Ferrichrome-iron receptor, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Interactions of TonB-dependent transporter FoxA with siderophores and antibiotics that affect binding, uptake, and signal transduction.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B8E
 
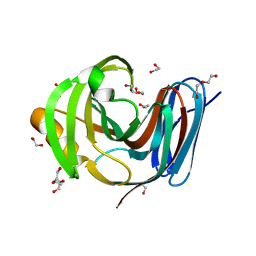 | | Wild-type GH11 from Blastobotrys mokoenaii | | Descriptor: | 1,2-ETHANEDIOL, BmGH11, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Coleman, T, Ravn, J.L, Larsbrink, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Yeasts Have Evolved Divergent Enzyme Strategies To Deconstruct and Metabolize Xylan.
Microbiol Spectr, 11, 2023
|
|
9HK5
 
 | | Structure of a mutant of human protein kinase CK2alpha' that equals its isoenzyme CK2alpha in affinity to the regulatory subunit CK2beta | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha' | | Authors: | Werner, C, Niefind, K. | | Deposit date: | 2024-12-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | A CK2 alpha ' mutant indicating why CK2 alpha and CK2 alpha ', the isoforms of the catalytic subunit of human protein kinase CK2, deviate in affinity to CK2 beta.
Biol.Chem., 406, 2025
|
|
6QZU
 
 | | Getah virus macro domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
9B2L
 
 | | Structure of the quorum quenching lactonase GcL D122N mutant - bimetallic center | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
7NBG
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z52314092, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBH
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z26781964, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
4X13
 
 | | JC Polyomavirus genotype 3 VP1 in complex with LSTc pentasaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
6R0P
 
 | | Getah virus macro domain in complex with ADPr in double open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Sulzenbacher, G, Ferreira Ramos, A.S, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
6R0T
 
 | | Getah virus macro domain in complex with ADPr in open conformation | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Non-structural polyprotein, ... | | Authors: | Ferreira Ramos, A.S, Sulzenbacher, G, Coutard, B. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Snapshots of ADP-ribose bound to Getah virus macro domain reveal an intriguing choreography.
Sci Rep, 10, 2020
|
|
