4XXL
 
 | |
3KQN
 
 | |
3KQU
 
 | |
1IVT
 
 | | NMR structures of the C-terminal globular domain of human lamin A/C | | Descriptor: | Lamin A/C | | Authors: | Krimm, I, Ostlund, C, Gilquin, B, Couprie, J, Hossenlopp, P, Mornon, J.P, Bonn, G, Courvalin, J.C, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy.
Structure, 10, 2002
|
|
2BRL
 
 | |
1JCI
 
 | | Stabilization of the Engineered Cation-binding Loop in Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome C Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Bonagura, C.A, Li, H, Poulos, T.L. | | Deposit date: | 2001-06-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cation-induced stabilization of the engineered cation-binding loop in cytochrome c peroxidase (CcP).
Biochemistry, 41, 2002
|
|
2BRK
 
 | |
5UQZ
 
 | | Structural Analysis of the Glucan Binding Protein C of Streptococcus mutans Provides Evidence that it Mediates both Sucrose-Independent and -Dependent Adherence | | Descriptor: | CALCIUM ION, Glucan-binding protein C, GbpC | | Authors: | Larson, M.R, Purushotham, S, Mieher, J, Wu, R, Rajashankar, K.R, Wu, H, Deivanayagam, C. | | Deposit date: | 2017-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Glucan Binding Protein C of Streptococcus mutans Mediates both Sucrose-Independent and Sucrose-Dependent Adherence.
Infect. Immun., 86, 2018
|
|
1W2A
 
 | | Deacetoxycephalosporin C synthase (with his-tag) complexed with Fe(II) and ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
1W28
 
 | | Conformational flexibility of the C-terminus with implications for substrate binding and catalysis in a new crystal form of deacetoxycephalosporin C synthase | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
1RWJ
 
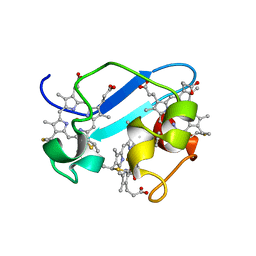 | | c7-type three-heme cytochrome domain | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Londer, Y.Y, Duke, N.E.C, Erickson, J, Pessanha, M, Salgueiro, C.A, Schiffer, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-08-03 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a novel c7-type three-heme cytochrome domain from a multidomain cytochrome c polymer.
Protein Sci., 13, 2004
|
|
5Y6G
 
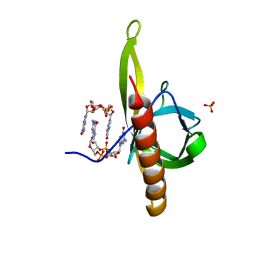 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
1I52
 
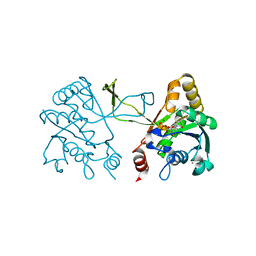 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHASE, CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, M, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-02-23 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1YWM
 
 | | Crystal structure of the N-terminal domain of group B Streptococcus alpha C protein | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C protein alpha-antigen, GLYCEROL | | Authors: | Auperin, T.C, Bolduc, G.R, Baron, M.J, Heroux, A, Filman, D.J, Madoff, L.C, Hogle, J.M. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the N-terminal domain of the group B streptococcus alpha C protein.
J.Biol.Chem., 280, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
3KQH
 
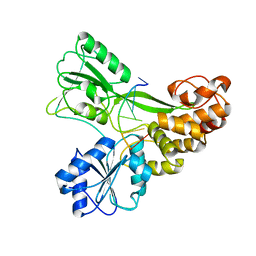 | |
3KQL
 
 | |
2BYF
 
 | | NMR solution structure of phospholipase c epsilon RA 2 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BYE
 
 | | NMR solution structure of phospholipase c epsilon RA 1 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
3FYE
 
 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides in the reduced state | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Qin, L, Mills, D.A, Proshlyakov, D.A, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Biochemistry, 48, 2009
|
|
2X07
 
 | | cytochrome c peroxidase: engineered ascorbate binding site | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
3L9H
 
 | | X-ray structure of mitotic kinesin-5 (KSP, KIF11, Eg5)in complex with the hexahydro-2H-pyrano[3,2-c]quinoline EMD 534085 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-{[(2R,4aS,5R,10bS)-5-phenyl-9-(trifluoromethyl)-3,4,4a,5,6,10b-hexahydro-2H-pyrano[3,2-c]quinolin-2-yl]methyl}urea, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11 | | Authors: | Knoechel, T. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery and optimization of hexahydro-2H-pyrano[3,2-c]quinolines (HHPQs) as potent and selective inhibitors of the mitotic kinesin-5.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
