7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | Descriptor: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
7LG4
 
 | |
7L31
 
 | | Cyro-EM structure of human Glycine Receptor alpha2-beta heteromer, strychnine bound state, 3.8 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-2, Glycine receptor subunit beta,Green fluorescent protein, ... | | Authors: | Yu, H, Wang, W. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Characterization of the subunit composition and structure of adult human glycine receptors
Neuron, 109, 2021
|
|
7KVV
 
 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, Squash RNA aptamer bound to DFHO | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVU
 
 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVT
 
 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T with iridium (III) ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KUY
 
 | |
7KS3
 
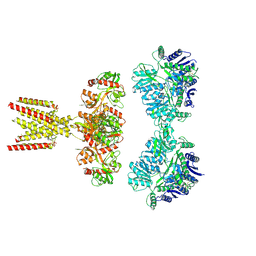 | | GluK2/K5 with L-Glu | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
7KS0
 
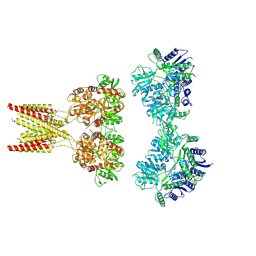 | | GluK2/K5 with 6-Cyano-7-nitroquinoxaline-2,3-dione (CNQX) | | Descriptor: | Glutamate receptor ionotropic, kainate 2, kainate 5,Green fluorescent protein chimera | | Authors: | Khanra, N, Brown, P.M.G.E, Perozzo, A.M, Bowie, D, Meyerson, J.R. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Architecture and structural dynamics of the heteromeric GluK2/K5 kainate receptor.
Elife, 10, 2021
|
|
7KM4
 
 | |
7KB5
 
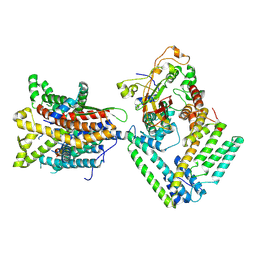 | |
7KAU
 
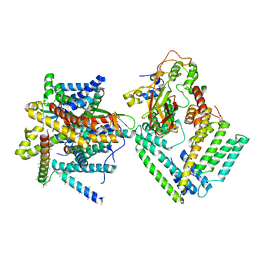 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAT
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAS
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAR
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec63 FN3 mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAQ
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class with Sec62, conformation 2 (C2) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAP
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAO
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAJ
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 2 (C2) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAI
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAH
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class without Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K18
 
 | | Cardiac Sodium channel with toxin bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-like toxin Lqh3, ... | | Authors: | Jiang, D, Catterall, W.A. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for voltage-sensor trapping of the cardiac sodium channel by a deathstalker scorpion toxin.
Nat Commun, 12, 2021
|
|
7JOT
 
 | |
7GQU
 
 | | Crystal Structure of Werner helicase fragment 517-945 in covalent complex with N-[(E,1S)-1-cyclopropyl-3-methylsulfonylprop-2-enyl]-2-(1,1-difluoroethyl)-4-phenoxypyrimidine-5-carboxamide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, GLYCEROL, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Tagliente, O, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
7GQT
 
 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, MAGNESIUM ION, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
