5FA7
 
 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
6EV0
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (AC129) | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, [(3~{R})-3-carboxy-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridin-8-yl]-dimethyl-azanium, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Cairns, A.G, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
8PZB
 
 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 49 | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
6EXK
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - unfolded HTH motif | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, SODIUM ION, ... | | Authors: | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
1BZO
 
 | | THREE-DIMENSIONAL STRUCTURE OF PROKARYOTIC CU,ZN SUPEROXIDE DISMUTASE FROM P.LEIOGNATHI, SOLVED BY X-RAY CRYSTALLOGRAPHY. | | Descriptor: | COPPER (II) ION, PROTEIN (SUPEROXIDE DISMUTASE), URANYL (VI) ION, ... | | Authors: | Bordo, D, Matak, D, Djinovic-Carugo, K, Rosano, C, Pesce, A, Bolognesi, M, Stroppolo, M.E, Falconi, M, Battistoni, A, Desideri, A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase.
J.Mol.Biol., 285, 1999
|
|
8PZ7
 
 | | crystal structure of VDR complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 57 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{R},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,8,9,9~{a}-hexahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-methyl-cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
1OE6
 
 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 5'-D(*CP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*G)-3', 5'-D(*CP*GP*GP*AP*CP*TP*3DRP*AP*CP*GP*GP*G)-3', 5-HYDROXYMETHYL URACIL, ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
8PZ9
 
 | | Crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 55 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{R},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,8,9,9~{a}-hexahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
1BZ4
 
 | | APOLIPOPROTEIN E3 (APO-E3), TRUNCATION MUTANT 165 | | Descriptor: | PROTEIN (APOLIPOPROTEIN E) | | Authors: | Rupp, B, Segelke, B. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational flexibility in the apolipoprotein E amino-terminal domain structure determined from three new crystal forms: implications for lipid binding.
Protein Sci., 9, 2000
|
|
1BXO
 
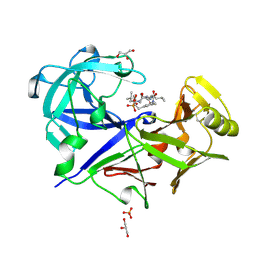 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
6OZW
 
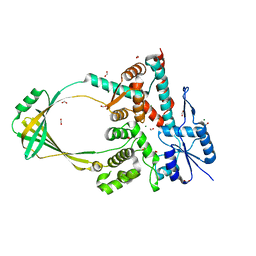 | |
8DD0
 
 | |
3IXX
 
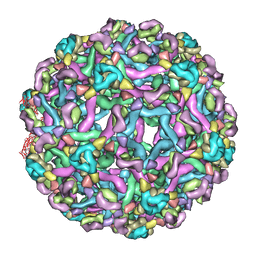 | | The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
7ZI4
 
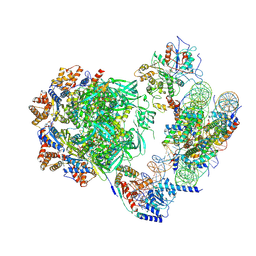 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
7SX6
 
 | |
7SX7
 
 | |
4XTJ
 
 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from 100 mM KCl plus 100 mM NaCl condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2NU6
 
 | | C123aA Mutant of E. coli Succinyl-CoA Synthetase | | Descriptor: | COENZYME A, SULFATE ION, Succinyl-CoA ligase [ADP-forming] subunit alpha, ... | | Authors: | Fraser, M.E. | | Deposit date: | 2006-11-08 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Participation of Cys 123alpha of Escherichia coli Succinyl-CoA Synthetase in Catalysis
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
1BB6
 
 | |
8CXR
 
 | | Crystal structure of MraY bound to a sphaerimicin analogue | | Descriptor: | (1S,4R,5S,6R,7S,9S,10S,11S,13S,14R)-9-[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]-14-(hexadecanoyloxy)-5,6,13-trihydroxy-8,16-dioxa-2,11-diazatricyclo[9.3.1.1~4,7~]hexadecane-10-carboxylic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2022-05-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Synthesis of macrocyclic nucleoside antibacterials and their interactions with MraY.
Nat Commun, 13, 2022
|
|
6BFQ
 
 | | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies | | Descriptor: | Fab Heavy chain, Fab Light Chain, Granulocyte-macrophage colony-stimulating factor | | Authors: | Dhagat, U, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2017-10-26 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies suggests novel therapeutic opportunities.
MAbs, 10, 2018
|
|
4XXJ
 
 | | Crystal Structure of Escherichia coli-Expressed Halobacterium salinarum Bacteriorhodopsin in the Trimeric Form | | Descriptor: | Bacteriorhodopsin, EICOSANE, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Bratanov, D, Balandin, T, Round, E, Gushchin, I, Gordeliy, V. | | Deposit date: | 2015-01-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Approach to Heterologous Expression of Membrane Proteins. The Case of Bacteriorhodopsin.
Plos One, 10, 2015
|
|
6R7P
 
 | | Crystal structure of oxidized Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF S96M | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gnandt, E, Friedrich, T. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
1ZG5
 
 | | NarL complexed to narG-89 promoter palindromic tail-to-tail DNA site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*CP*TP*AP*TP*AP*GP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/nitrite response regulator protein narL, SULFATE ION | | Authors: | Maris, A.E, Kaczor-Grzeskowiak, M, Ma, Z, Kopka, M.L, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2005-04-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Biochemistry, 44, 2005
|
|
1O9R
 
 | | The X-ray crystal structure of Agrobacterium tumefaciens Dps, a member of the family that protect DNA without binding | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGROBACTERIUM TUMEFACIENS DPS, ... | | Authors: | Ilari, A, Ceci, P, Chiancone, E. | | Deposit date: | 2002-12-18 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Dps Protein of Agrobacterium Tumefaciens Does not Bind to DNA But Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties
J.Biol.Chem., 278, 2003
|
|
