5SRZ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5281440906 - (R,S) and (S,R) isomers | | Descriptor: | (1R,2S)-2-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclohexane-1-carboxylic acid, (1S,2R)-2-[(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)amino]cyclohexane-1-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ5
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894407 - (R,S) and (S,S) isomers | | Descriptor: | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5L1U
 
 | |
5SPN
 
 | |
5SPM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with FRESH00002410346 | | Descriptor: | 4-hydroxy-6-(3-hydroxy-1-methyl-1,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridine-6-carbonyl)-2H-pyran-2-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPO
 
 | |
5SPP
 
 | |
5SPR
 
 | |
5SPU
 
 | |
5SPT
 
 | |
5SPS
 
 | |
5SPV
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with REAL250003774401 | | Descriptor: | 2-[methyl-[(9-oxidanylidene-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl)carbonyl]amino]ethanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPQ
 
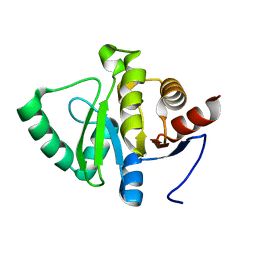 | |
5SPW
 
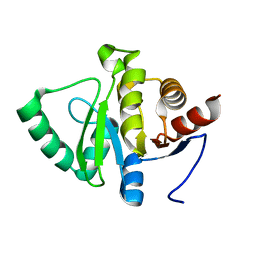 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with FRESH00004674769 - (R,S,R) isomer | | Descriptor: | (1R,5S,6R)-3-(7H-purin-6-yl)-3-azabicyclo[3.2.2]nonane-6-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5L1T
 
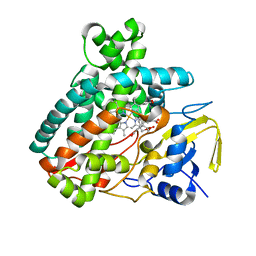 | |
2DYH
 
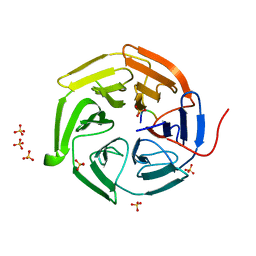 | |
4URR
 
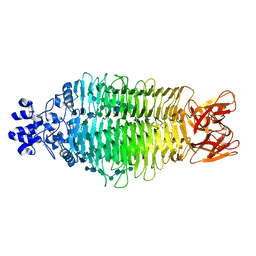 | | Tailspike protein of Sf6 bacteriophage bound to Shigella flexneri O- antigen octasaccharide fragment | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL TAIL PROTEIN, MANGANESE (II) ION, ... | | Authors: | Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2014-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacteriophage Tailspikes and Bacterial O-Antigens as a Model System to Study Weak-Affinity Protein-Polysaccharide Interactions.
J.Am.Chem.Soc., 138, 2016
|
|
1M1U
 
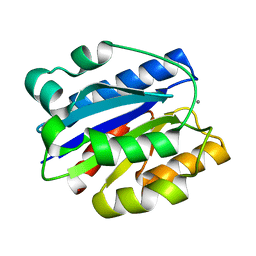 | | AN ISOLEUCINE-BASED ALLOSTERIC SWITCH CONTROLS AFFINITY AND SHAPE SHIFTING IN INTEGRIN CD11B A-DOMAIN | | Descriptor: | CALCIUM ION, Integrin alpha-M | | Authors: | Xiong, J.-P, Li, R, Essafi, M, Stehle, T, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain.
J.Biol.Chem., 275, 2000
|
|
3TDI
 
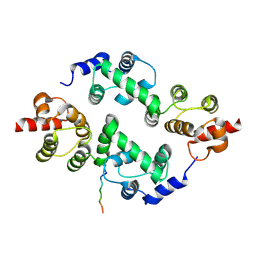 | | yeast Cul1WHB-Dcn1P acetylated Ubc12N complex | | Descriptor: | Defective in cullin neddylation protein 1, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
1BSO
 
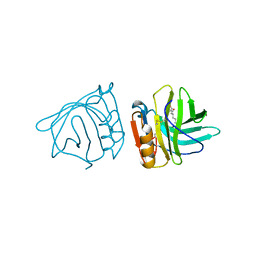 | | 12-BROMODODECANOIC ACID BINDS INSIDE THE CALYX OF BOVINE BETA-LACTOGLOBULIN | | Descriptor: | 12-BROMODODECANOIC ACID, PROTEIN (BOVINE BETA-LACTOGLOBULIN A) | | Authors: | Qin, B.Y, Creamer, L.K, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | 12-Bromododecanoic acid binds inside the calyx of bovine beta-lactoglobulin.
FEBS Lett., 438, 1998
|
|
4W8O
 
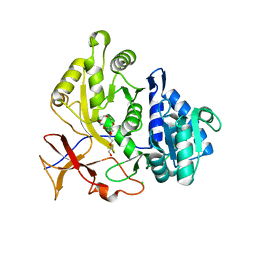 | | Structure of the luciferase-like enzyme from the nonluminescent Zophobas morio mealworm | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, luciferase-like enzymeAMP-CoA-ligase | | Authors: | Santos, C.R, Prado, R.A, Viviani, V, Murakami, M.T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the luciferase-like enzyme from the nonluminescent Zophobas morio mealworm
To Be Published
|
|
1MX9
 
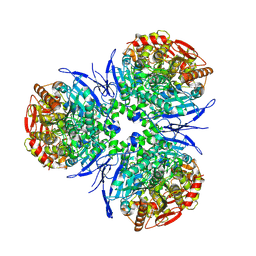 | | Crystal Structure of Human Liver Carboxylesterase in complexed with naloxone methiodide, a heroin analogue | | Descriptor: | (5A,17R)-4,5-EPOXY-3,14-DIHYDROXY-17-METHYL-6-OXO-17-(2-PROPENYL)-MORPHINANIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, liver Carboxylesterase I | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
1NJ1
 
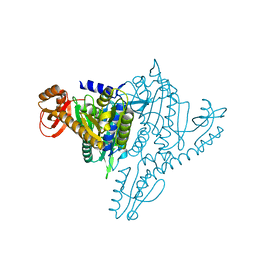 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to cysteine sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1AH7
 
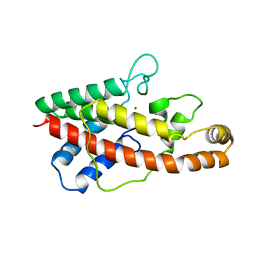 | | PHOSPHOLIPASE C FROM BACILLUS CEREUS | | Descriptor: | PHOSPHOLIPASE C, ZINC ION | | Authors: | Greaves, R. | | Deposit date: | 1997-04-14 | | Release date: | 1997-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus.
Nature, 338, 1989
|
|
7SJQ
 
 | |
