1SDV
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
1SDW
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase with bound peptide and dioxygen | | Descriptor: | COPPER (II) ION, GLYCEROL, N-ALPHA-ACETYL-3,5-DIIODOTYROSYL-D-THREONINE, ... | | Authors: | Prigge, S.T, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dioxygen binds end-on to mononuclear copper in a precatalytic enzyme complex.
Science, 304, 2004
|
|
1SDX
 
 | | Crystal structure of the zinc saturated C-terminal half of bovine lactoferrin at 2.0 A resolution reveals two additional zinc binding sites | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, Lactotransferrin, ... | | Authors: | Jabeen, T, Sharma, S, Singhal, G, Singh, N, Singh, T.P. | | Deposit date: | 2004-02-15 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of the zinc-saturated C-terminal lobe of bovine lactoferrin at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SDY
 
 | | STRUCTURE SOLUTION AND MOLECULAR DYNAMICS REFINEMENT OF THE YEAST CU,ZN ENZYME SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic, K, Gatti, G, Coda, A, Antolini, L, Pelosi, G, Desideri, A, Falconi, M, Marmocchi, F, Rotilio, G, Bolognesi, M. | | Deposit date: | 1991-06-14 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure solution and molecular dynamics refinement of the yeast Cu,Zn enzyme superoxide dismutase.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1SDZ
 
 | | Crystal structure of DIAP1 BIR1 bound to a Reaper peptide | | Descriptor: | Apoptosis 1 inhibitor, Reaper, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SE0
 
 | | Crystal structure of DIAP1 BIR1 bound to a Grim peptide | | Descriptor: | Apoptosis 1 inhibitor, Cell death protein Grim, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SE2
 
 | |
1SE3
 
 | | STAPHYLOCOCCAL ENTEROTOXIN B COMPLEXED WITH GM3 TRISACCHARIDE | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, STAPHYLOCOCCAL ENTEROTOXIN B | | Authors: | Swaminathan, S, Sax, M. | | Deposit date: | 1996-10-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Residues defining V beta specificity in staphylococcal enterotoxins.
Nat.Struct.Biol., 2, 1995
|
|
1SE4
 
 | | STAPHYLOCOCCAL ENTEROTOXIN B COMPLEXED WITH LACTOSE | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN B, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Swaminathan, S, Sax, M. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Residues defining V beta specificity in staphylococcal enterotoxins.
Nat.Struct.Biol., 2, 1995
|
|
1SE6
 
 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SPERMINE (FULLY PROTONATED FORM), ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-02-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2.
J.Biol.Chem., 280, 2005
|
|
1SE7
 
 | | Solution structure of the E. coli bacteriophage P1 encoded HOT protein: a homologue of the theta subunit of E. coli DNA polymerase III | | Descriptor: | HOMOLOGUE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Chikova, A.K, Schaaper, R.M, London, R.E. | | Deposit date: | 2004-02-16 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage Like It HOT: Solution Structure of the Bacteriophage P1-Encoded HOT Protein, a Homolog of the theta Subunit of E. coli DNA Polymerase III
Structure, 12, 2004
|
|
1SE8
 
 | | Structure of single-stranded DNA-binding protein (SSB) from D. radiodurans | | Descriptor: | Single-strand binding protein | | Authors: | Bernstein, D.A, Eggington, J.M, Killoran, M.P, Misic, A.M, Cox, M.M, Keck, J.L. | | Deposit date: | 2004-02-16 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SE9
 
 | |
1SEB
 
 | | COMPLEX OF THE HUMAN MHC CLASS II GLYCOPROTEIN HLA-DR1 AND THE BACTERIAL SUPERANTIGEN SEB | | Descriptor: | ENDOGENOUS PEPTIDE MODEL, POLY-ALA, ENTEROTOXIN TYPE B, ... | | Authors: | Jardetzky, T.S, Brown, J.H, Gorga, J.C, Stern, L.J, Urban, R.G, Chi, Y.I, Stauffacher, C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1995-11-26 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen.
Nature, 368, 1994
|
|
1SED
 
 | | Crystal Structure of Protein of Unknown Function YhaL from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Hypothetical protein yhaI, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Hypothetical Protein YhaI, APC1180 from Bacillus subtilis
To be Published
|
|
1SEF
 
 | |
1SEG
 
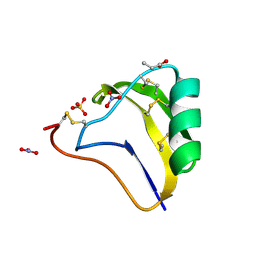 | | Crystal structure of a toxin chimera between Lqh-alpha-IT from the scorpion Leiurus quinquestriatus hebraeus and AAH2 from Androctonus australis hector | | Descriptor: | AAH2: LQH-ALPHA-IT (FACE) CHIMERIC TOXIN, NITRATE ION, PROPANOIC ACID, ... | | Authors: | Karbat, I, Frolow, F, Froy, O, Gilles, N, Cohen, L, Turkov, M, Gordon, D, Gurevitz, M. | | Deposit date: | 2004-02-17 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of the high insecticidal potency of scorpion alpha-toxins.
J.Biol.Chem., 279, 2004
|
|
1SEH
 
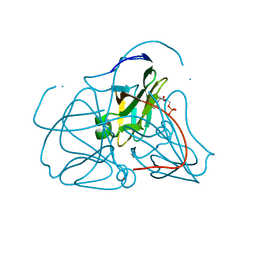 | | Crystal structure of E. coli dUTPase complexed with the product dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-02-17 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
1SEI
 
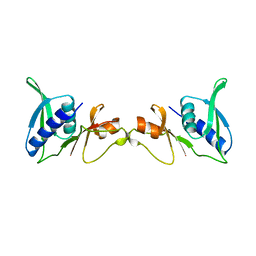 | | STRUCTURE OF 30S RIBOSOMAL PROTEIN S8 | | Descriptor: | RIBOSOMAL PROTEIN S8 | | Authors: | Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1996-08-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence for specific S8-RNA and S8-protein interactions within the 30S ribosomal subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 A resolution.
Structure, 4, 1996
|
|
1SEJ
 
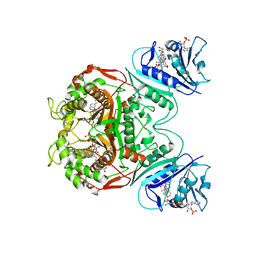 | | Crystal Structure of Dihydrofolate Reductase-Thymidylate Synthase from Cryptosporidium hominis Bound to 1843U89/NADPH/dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, ... | | Authors: | Anderson, A.C. | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Two crystal structures of dihydrofolate reductase-thymidylate synthase from Cryptosporidium hominis reveal protein-ligand interactions including a structural basis for observed antifolate resistance.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1SEK
 
 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | Descriptor: | SERPIN K | | Authors: | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
1SEL
 
 | |
1SEM
 
 | |
1SEN
 
 | | Endoplasmic reticulum protein Rp19 O95881 | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Thioredoxin-like protein p19 | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Lee, D, Dailey, T.A, Mayer, M.R, Rose, J.P, Richardson, D.C, Richardson, J.S, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-07-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Endoplasmic reticulum protein Rp19
To be Published
|
|
1SEP
 
 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND SEPIAPTERIN | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SEPIAPTERIN REDUCTASE | | Authors: | Auerbach, G, Herrmann, A, Guetlich, M, Fischer, M, Jacob, U, Bacher, A, Huber, R. | | Deposit date: | 1997-05-23 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
