5WB0
 
 | |
4N9Y
 
 | |
4N9L
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
6SW1
 
 | | Crystal Structure of P. aeruginosa PqsL: R41Y, I43R, G45R, C105G mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Mattevi, A, Rovida, S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoinduced monooxygenation involving NAD(P)H-FAD sequential single-electron transfer.
Nat Commun, 11, 2020
|
|
4NA0
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4NAY
 
 | | Crystal Structure of FosB from Staphylococcus aureus with Zn and Sulfate at 1.42 Angstrom Resolution - SAD Phasing | | Descriptor: | Metallothiol transferase FosB, SULFATE ION, ZINC ION | | Authors: | Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Cook, P.D, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
5WDJ
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH COMPOUND-6 AKA 7-(BENZYLOXY)-1H-[1,2, 3]TRIAZOLO[4,5-D]PYRIMIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(benzyloxy)-1H-[1,2,3]triazolo[4,5-d]pyrimidin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2017-07-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Triazolopyrimidines identified as reversible myeloperoxidase inhibitors.
Medchemcomm, 8, 2017
|
|
4NB2
 
 | | Crystal Structure of FosB from Staphylococcus aureus at 1.89 Angstrom Resolution - Apo structure | | Descriptor: | Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
6SMS
 
 | | Vegetative Insecticidal Protein 1 (Vip1Ac1) from Bacillus thuringiensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Rizkallah, P.J, Al-Maslookhi, H.S, Berry, C, Jones, D. | | Deposit date: | 2019-08-22 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bacillus thuringiensis insecticidal protein 1Ac, VIP1Ac
Not Published
|
|
6SM0
 
 | | Venus 66 p-Azido-L-Phenylalanin (azF) variant, dark grown | | Descriptor: | Green fluorescent protein, OXYGEN MOLECULE, SULFATE ION, ... | | Authors: | Rizkallah, P.J, Al Maslookhi, H.S, Jones, D.D. | | Deposit date: | 2019-08-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Stalling chromophore synthesis of the fluorescent protein Venus reveals the molecular basis of the final oxidation step.
Chem Sci, 12, 2021
|
|
4NBR
 
 | |
4MPW
 
 | | Human beta-tryptase co-crystal structure with [(1,1,3,3-tetramethyldisiloxane-1,3-diyl)di-1-benzofuran-3,5-diyl]bis({4-[3-(aminomethyl)phenyl]piperidin-1-yl}methanone) | | Descriptor: | ACETATE ION, CHLORIDE ION, TRIETHYLENE GLYCOL, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel, Nonpeptidic, Orally Active Bivalent Inhibitor of Human beta-Tryptase.
Pharmacology, 102, 2018
|
|
4MP6
 
 | | Staphyloferrin B precursor biosynthetic enzyme SbnB bound to citrate and NAD+ | | Descriptor: | CITRIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative ornithine cyclodeaminase | | Authors: | Grigg, J.C, Kobylarz, M.J, Rai, D.K, Murphy, M.E.P. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis of L-2,3-diaminopropionic Acid, a siderophore and antibiotic precursor.
Chem.Biol., 21, 2014
|
|
4MPX
 
 | | Human beta-tryptase co-crystal structure with [(1,1,3,3-tetramethyldisiloxane-1,3-diyl)di-1-benzothiene-4,2-diyl]bis({4-[3-(aminomethyl)phenyl]piperidin-1-yl}methanone) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Target-directed self-assembly of homodimeric drugs
To be Published
|
|
4MQA
 
 | | Human beta-tryptase co-crystal structure with {(1,1,3,3-tetramethyldisiloxane-1,3-diyl)bis[5-(methylsulfanyl)benzene-3,1-diyl]}bis({4-[3-(aminomethyl)phenyl]piperidin-1-yl}methanone) | | Descriptor: | SULFATE ION, TETRAETHYLENE GLYCOL, Tryptase alpha/beta-1, ... | | Authors: | White, A, Lakshminarasimhan, D, Suto, R. | | Deposit date: | 2013-09-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Target-directed self-assembly of homodimeric drugs
To be Published
|
|
6STV
 
 | | Adenovirus 29 Fiber Knob protein | | Descriptor: | Fiber, GLYCEROL, PHOSPHATE ION | | Authors: | Mundy, R.M, Baker, A.T, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2019-09-11 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus
npj Viruses, 1, 2023
|
|
4MQJ
 
 | |
4MRG
 
 | | Crystal structure of the murine cd44 hyaluronan binding domain complex with a small molecule | | Descriptor: | 1,2,3,4-tetrahydroisoquinolin-5-amine, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Liu, L.K, Finzel, B. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-Based Identification of an Inducible Binding Site on Cell Surface Receptor CD44 for the Design of Protein-Carbohydrate Interaction Inhibitors.
J.Med.Chem., 57, 2014
|
|
4MRQ
 
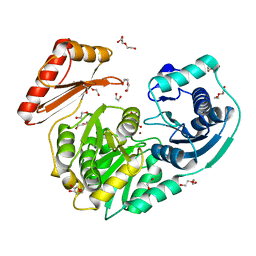 | | Crystal Structure of wild-type unphosphorylated PMM/PGM | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, ... | | Authors: | Lee, Y, Beamer, L. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase.
J.Biol.Chem., 289, 2014
|
|
6STW
 
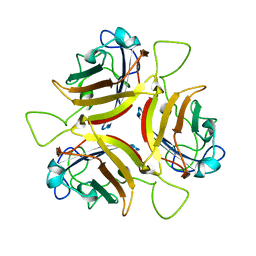 | |
5WDE
 
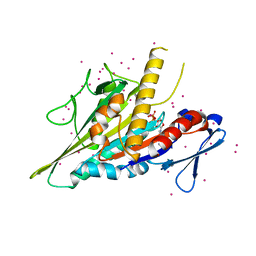 | | Crystal structure of the KIFC3 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC3, MAGNESIUM ION, ... | | Authors: | Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
4MT1
 
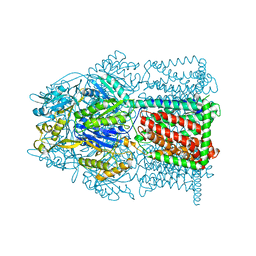 | |
4MSO
 
 | |
4MTL
 
 | | Human Methyltransferase-Like Protein 21C | | Descriptor: | Protein-lysine methyltransferase METTL21C, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | Hong, B.S, Tempel, W, Dong, A, Li, Y, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Human Methyltransferase-Like Protein 21C
To be Published
|
|
4V6F
 
 | | Elongation complex of the 70S ribosome with three tRNAs and mRNA. | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 23S RRNA, ... | | Authors: | Jenner, L.B, Yusupova, G, Yusupov, M. | | Deposit date: | 2009-07-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural aspects of messenger RNA reading frame maintenance by the ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
