9ER3
 
 | |
8Z8S
 
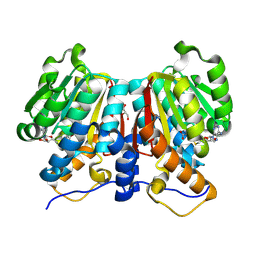 | |
8VAN
 
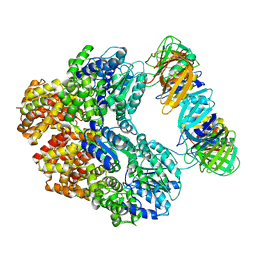 | | Structure of the E. coli clamp loader bound to the beta clamp in an Initial-Binding conformation | | Descriptor: | Beta sliding clamp, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Landeck, J.T, Pajak, J, Kelch, B.A. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Differences between bacteria and eukaryotes in clamp loader mechanism, a conserved process underlying DNA replication.
J.Biol.Chem., 300, 2024
|
|
8XEG
 
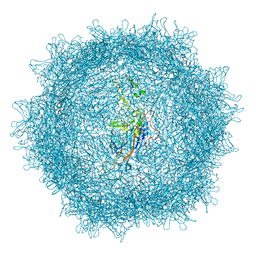 | |
8W56
 
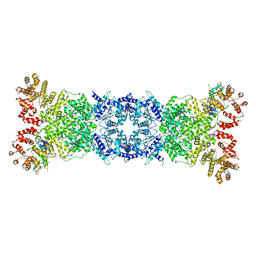 | | Cryo-EM structure of DSR2-DSAD1 state 1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-08-25 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8W1S
 
 | | Cryo-EM structure of BTV pre-core | | Descriptor: | Core protein VP3, RNA-directed RNA polymerase | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8Y4B
 
 | |
8VD1
 
 | | Crystal Structure of KAI2 from Oryza sativa with MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Probable esterase D14L | | Authors: | Gilio, A.K, Shabek, N, Guercio, A.M, Pawlak, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insights into rice KAI2 receptor provide functional implications for perception and signal transduction.
J.Biol.Chem., 300, 2024
|
|
9C0Y
 
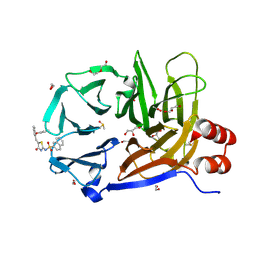 | | Clathrin terminal domain complexed with Pitstop 2c | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Clathrin heavy chain 1, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
8VEU
 
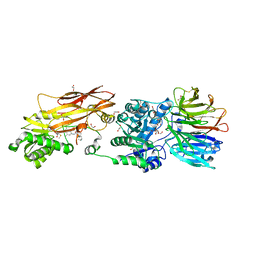 | |
9BOM
 
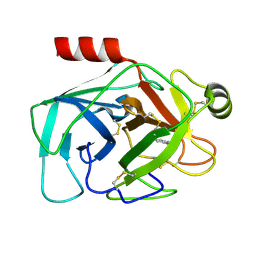 | | Crystal structure of reduced bovine trypsin, 25mM DTT-treated | | Descriptor: | BENZAMIDINE, Cationic trypsin | | Authors: | Zhou, D, Chen, L, Rose, J.P, Wang, B.C. | | Deposit date: | 2024-05-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of reduced bovine trypsin, 25mM DTT-treated
To Be Published
|
|
8XU4
 
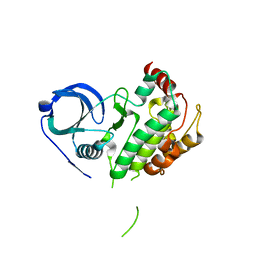 | | The Crystal Structure of MAPK2 from Biortus. | | Descriptor: | MALONIC ACID, MAP kinase-activated protein kinase 2 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-01-12 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Crystal Structure of MAPK2 from Biortus.
To Be Published
|
|
9EWQ
 
 | |
8XOY
 
 | | The Crystal Structure of PTP1B from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Li, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of PTP1B from Biortus.
To Be Published
|
|
8X9E
 
 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in low PEG concentration | | Descriptor: | 1,2-ETHANEDIOL, Carbon monoxide dehydrogenase 2, FE (III) ION, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8VET
 
 | |
8YGW
 
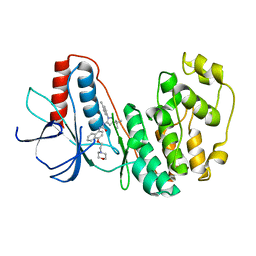 | | The Crystal Structure of MAPK11 from Biortus | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The Crystal Structure of MAPK11 from Biortus.
To Be Published
|
|
9C1Q
 
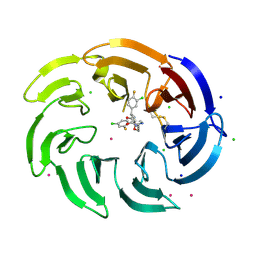 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-39512 compound | | Descriptor: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-(3-methoxyprop-1-yn-1-yl)-1H-pyrrole-3-carboxamide, CHLORIDE ION, DDB1- and CUL4-associated factor 1, ... | | Authors: | kimani, S, Dong, A, Li, Y, Seitova, A, Al-awar, R, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-05-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-39512 compound
To be published
|
|
8Y74
 
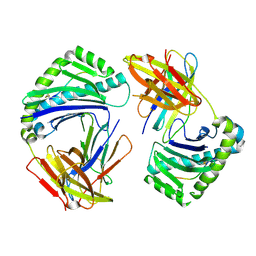 | |
8W5C
 
 | | Crystal structure of FGFR4 kinase domain in complex with 8K | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-6-[(4-methylpiperazin-1-yl)methyl]-1-propan-2-yl-pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Zhang, Z.M, Huang, H.S. | | Deposit date: | 2023-08-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 6-Formylpyridyl Urea Derivatives as Potent Reversible-Covalent Fibroblast Growth Factor Receptor 4 Inhibitors with Improved Anti-Hepatocellular Carcinoma Activity.
J.Med.Chem., 67, 2024
|
|
8YOO
 
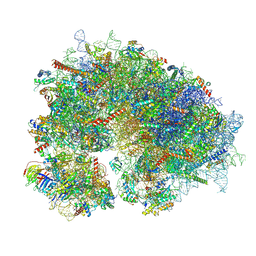 | | Cryo-EM structure of the human 80S ribosome with 100 um Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Denk, T, Cheng, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
9C11
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36R at cryogenic temperature | | Descriptor: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain-swapping promoted by the introduction of a charge in the hydrophobic interior of a protein
To Be Published
|
|
8YBB
 
 | |
8Y6Q
 
 | | Structure of the Dark/Dronc complex | | Descriptor: | Apaf-1 related killer DARK, Caspase Dronc | | Authors: | Tian, L, Li, Y, Shi, Y. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Dark and Dronc activation in Drosophila melanogaster.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8W9Q
 
 | | Structure of partial Banna virus | | Descriptor: | VP10, VP2, VP4, ... | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-09-05 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
