2N17
 
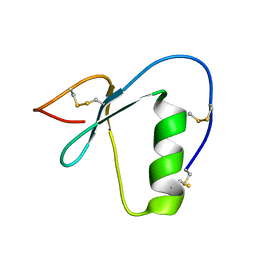 | | NMR structure of a Kazal-type serine protease inhibitor from the subterranean termite defense gland of Coptotermes formosanus Shiraki soldiers | | Descriptor: | Lysozyme-Protease Inhibitor Protein | | Authors: | Negulescu, H, Guo, Y, Garner, T.P, Goodwin, O.Y, Henderson, G, Laine, R.A, Macnaughtan, M.A. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Kazal-Type Serine Protease Inhibitor from the Defense Gland Secretion of the Subterranean Termite Coptotermes formosanus Shiraki.
Plos One, 10, 2015
|
|
2AAV
 
 | | Solution NMR structure of Filamin A domain 17 | | Descriptor: | Filamin A | | Authors: | Nakamura, F, Pudas, R, Heikkinen, O, Permi, P, Kilpelainen, I, Munday, A.D, Hartwig, J.H, Stossel, T.P, Ylanne, J. | | Deposit date: | 2005-07-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the GPIb-filamin A complex
Blood, 107, 2006
|
|
4CP9
 
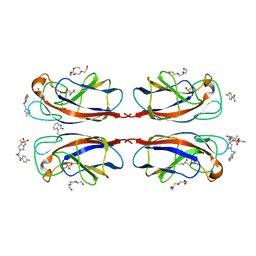 | | Crystal structure OF lecA lectin complexed with a divalent galactoside at 1.65 angstrom | | Descriptor: | (4S)-N-ethyl-4-{[N-methyl-3-(1-{2-[(4-sulfanylbenzoyl)amino]ethyl}-1H-1,2,3-triazol-4-yl)-L-alanyl]amino}-L-prolinamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Topin, J, Varrot, A, Imberty, A, Wissinger, N. | | Deposit date: | 2014-02-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Leca Ligand Identified from a Galactoside-Conjugate Array Inhibits Host Cell Invasion by Pseudomonas Aeruginosa.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1YBG
 
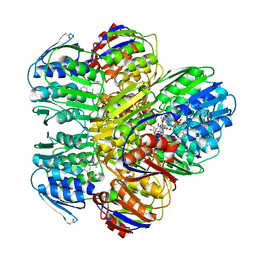 | | MurA inhibited by a derivative of 5-sulfonoxy-anthranilic acid | | Descriptor: | N-METHYL-N-{2-[(2-NAPHTHYLSULFONYL)AMINO]-5-[(2-NAPHTHYLSULFONYL)OXY]BENZOYL}-L-ASPARTIC ACID, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Eschenburg, S, Priestman, M.A, Abdul-Latif, F.A, Delachaume, C, Fassy, F, Schonbrunn, E. | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Inhibitor That Suspends the Induced Fit Mechanism of UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA).
J.Biol.Chem., 280, 2005
|
|
1KWJ
 
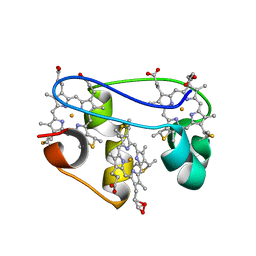 | | solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans, minimized average structure | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
2V8J
 
 | |
3RZK
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(EDA)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*TP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZJ
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(ME6)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
4E8F
 
 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | ACETATE ION, GLYCEROL, Poly(A) RNA polymerase protein cid1 | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1M4L
 
 | | STRUCTURE OF NATIVE CARBOXYPEPTIDASE A AT 1.25 RESOLUTION | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Kilshtain-Vardi, A, Glick, M, Greenblatt, H.M, Goldblum, A, Shoham, G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-01-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Refined structure of bovine carboxypeptidase A at 1.25 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4IAU
 
 | | Atomic resolution structure of Geodin, a beta-gamma crystallin from Geodia cydonium | | Descriptor: | Beta-gamma-crystallin, CALCIUM ION, GLYCEROL | | Authors: | Vergara, A, Grassi, M, Sica, F, Mazzarella, L, Merlino, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A novel interdomain interface in crystallins: structural characterization of the [beta][gamma]-crystallin from Geodia cydonium at 0.99 A resolution
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1EDI
 
 | |
1MTP
 
 | | The X-ray crystal structure of a serpin from a thermophilic prokaryote | | Descriptor: | Serine Proteinase Inhibitor (SERPIN), Chain A, Chain B | | Authors: | Irving, J.A, Cabrita, L.D, Rossjohn, J, Pike, R.N, Bottomley, S.P, Whisstock, J.C. | | Deposit date: | 2002-09-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Structure, 11, 2003
|
|
4EQW
 
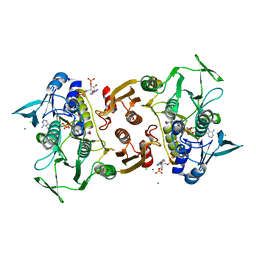 | | Crystal Structure of the Y361F, Y419F Mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Edwards, J.S, Wallace, B.D, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
1MUJ
 
 | |
1EDK
 
 | |
4EQS
 
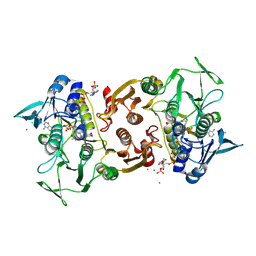 | | Crystal structure of the Y419F mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Wallace, B.D, Edwards, J.S, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4IN7
 
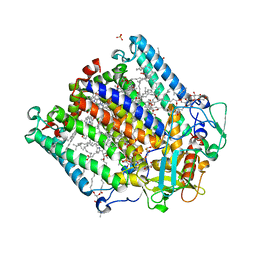 | | (M)L214N mutant of the Rhodobacter sphaeroides Reaction Center | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Saer, R.G, Hardjasa, A, Murphy, M.E.P, Beatty, J.T. | | Deposit date: | 2013-01-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Role of Rhodobacter sphaeroides photosynthetic reaction center residue M214 in the composition, absorbance properties, and conformations of H(A) and B(A) cofactors.
Biochemistry, 52, 2013
|
|
1HJ1
 
 | | RAT OESTROGEN RECEPTOR BETA LIGAND-BINDING DOMAIN IN COMPLEX WITH PURE ANTIOESTROGEN ICI164,384 | | Descriptor: | N-BUTYL-11-[(7R,8R,9S,13S,14S,17S)-3,17-DIHYDROXY-13-METHYL-7,8,9,11,12,13,14,15,16,17-DECAHYDRO-6H-CYCLOPENTA[A]PHENANTHREN-7-YL]-N-METHYLUNDECANAMIDE, NICKEL (II) ION, OESTROGEN RECEPTOR BETA, ... | | Authors: | Pike, A.C.W, Brzozowski, A.M, Carlquist, M. | | Deposit date: | 2001-01-08 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into the Mode of Action of a Pure Antiestrogen
Structure, 9, 2001
|
|
1NXD
 
 | | Crystal structure of MnMn Concanavalin A | | Descriptor: | AZIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lopez-Jaramillo, F.J, Gonzalez-Ramirez, L.A, Albert, A, Santoyo-Gonzalez, F, Vargas-Berenguel, A, Otalora, F. | | Deposit date: | 2003-02-10 | | Release date: | 2004-03-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of concanavalin A at pH 8: bound solvent and crystal contacts.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4JC3
 
 | |
1EDJ
 
 | |
1EDL
 
 | |
5RSA
 
 | |
1CCI
 
 | | HOW FLEXIBLE ARE PROTEINS? TRAPPING OF A FLEXIBLE LOOP | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1996-12-18 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
