6KWJ
 
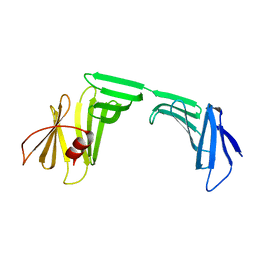 | | A Crystal Structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Kiya, M, Makabe, K. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | beta-Strand-mediated Domain-swapping in the Absence of Hydrophobic Core Repacking.
J.Mol.Biol., 436, 2024
|
|
6KWV
 
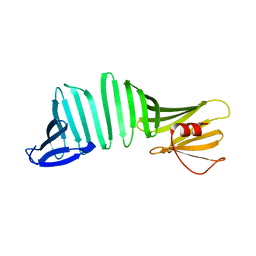 | | A Crystal Structure of OspA mutant | | Descriptor: | Outer surface protein A | | Authors: | Kiya, M, Makabe, K. | | Deposit date: | 2019-09-08 | | Release date: | 2020-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | beta-Strand-mediated Domain-swapping in the Absence of Hydrophobic Core Repacking.
J.Mol.Biol., 436, 2024
|
|
2OUI
 
 | | D275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Frolow, F, Shimon, L, Burstein, Y, Goihberg, E, Peretz, M, Dym, O. | | Deposit date: | 2007-02-11 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.
Proteins, 72, 2008
|
|
1UXL
 
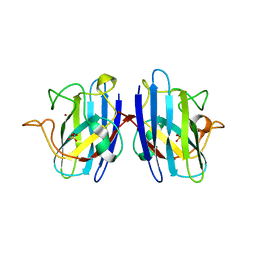 | | I113T mutant of human SOD1 | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Hough, M.A, Grossmann, J.G, Antonyuk, S.V, Strange, R.W, Doucette, P.A, Rodriguez, J.A, Whitson, L.J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2004-02-25 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimer Destabilization in Superoxide Dismutase May Result in Disease-Causing Properties: Structures of Motor Neuron Disease Mutants
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6KZ7
 
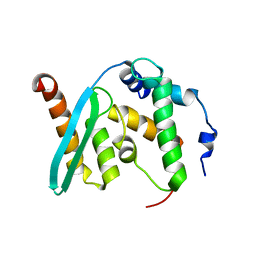 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
1BDM
 
 | |
6LJY
 
 | | A Crystal Structure of OspA mutant | | Descriptor: | OspA163 | | Authors: | Kiya, M, Makabe, K. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | beta-Strand-mediated Domain-swapping in the Absence of Hydrophobic Core Repacking.
J.Mol.Biol., 436, 2024
|
|
1UYV
 
 | |
6LEA
 
 | |
1UYT
 
 | | Acetyl-CoA carboxylase carboxyltransferase domain | | Descriptor: | ACETYL-COA CARBOXYLASE | | Authors: | Zhang, H, Tweel, B, Tong, L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Inhibition of the Carboxyltransferase Domain of Acetyl-Coenzyme-A Carboxylase by Haloxyfop and Diclofop.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6KSS
 
 | |
1SHY
 
 | |
1BGF
 
 | |
6KZQ
 
 | | structure of PTP-MEG2 and NSF-pY83 peptide complex | | Descriptor: | NSF-pY83 peptide, Tyrosine-protein phosphatase non-receptor type 9 | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
6KZA
 
 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | Descriptor: | DNA replication protein DnaC, Replicative DNA helicase | | Authors: | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
6L03
 
 | | structure of PTP-MEG2 and MUNC18-1-pY145 peptide complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 9, stxbp1-pY145 peptide | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
1BA9
 
 | | THE SOLUTION STRUCTURE OF REDUCED MONOMERIC SUPEROXIDE DISMUTASE, NMR, 36 STRUCTURES | | Descriptor: | COPPER (I) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Banci, L, Benedetto, M, Bertini, I, Del Conte, R, Piccioli, M, Viezzoli, M.S. | | Deposit date: | 1998-04-24 | | Release date: | 1998-09-16 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced monomeric Q133M2 copper, zinc superoxide dismutase (SOD). Why is SOD a dimeric enzyme?.
Biochemistry, 37, 1998
|
|
1SMB
 
 | | Crystal Structure of Golgi-Associated PR-1 protein | | Descriptor: | 17kD fetal brain protein | | Authors: | Serrano, R.L, Kuhn, A, Hendricks, A, Helms, J.B, Sinning, I, Groves, M.R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism
J.Mol.Biol., 339, 2004
|
|
2R27
 
 | | Constitutively zinc-deficient mutant of human superoxide dismutase (SOD), C6A, H80S, H83S, C111S | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn] | | Authors: | Roberts, B.R, Getzoff, E.D, Karplus, P.A, Beckman, J.S, Tainer, J.A. | | Deposit date: | 2007-08-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS.
J.Mol.Biol., 373, 2007
|
|
2R0Q
 
 | |
2A5S
 
 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | Descriptor: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2A5T
 
 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | Descriptor: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
1Y52
 
 | | structure of insect cell (Baculovirus) expressed AVR4 (C122S)-biotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin-related protein 4/5, BIOTIN | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Y7Q
 
 | | Mammalian SCAN domain dimer is a domain-swapped homologue of the HIV capsid C-terminal domain | | Descriptor: | Zinc finger protein 174 | | Authors: | Ivanov, D, Stone, J.R, Maki, J.L, Collins, T, Wagner, G. | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mammalian SCAN Domain Dimer Is a Domain-Swapped Homolog of the HIV Capsid C-Terminal Domain
Mol.Cell, 17, 2005
|
|
2LJY
 
 | | Haddock model structure of the N-terminal domain dimer of HPV16 E6 | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
