6KZU
 
 | |
8VJP
 
 | | Histidine-covalent stapled alpha-helical peptide (155H1) targeting hMcl-1 | | Descriptor: | (4Z)-oct-4-en-1-ol, (S~1~R)-3-carbamoyl-4-methoxybenzene-1-sulfinic acid, Histidine-covalent stapled alpha-helical peptide, ... | | Authors: | Muzzarelli, K.M, Assar, Z, Alboreggia, G, Pellecchia, M. | | Deposit date: | 2024-01-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Histidine-Covalent Stapled Alpha-Helical Peptides Targeting hMcl-1.
J.Med.Chem., 67, 2024
|
|
6L4T
 
 | | Structure of the peripheral FCPI from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
6L31
 
 | | L1 protein of human papillomavirus 6 | | Descriptor: | Major capsid protein L1 | | Authors: | Li, S.W, Liu, X.L, Gu, Y. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Neutralization sites of human papillomavirus-6 relate to virus attachment and entry phase in viral infection.
Emerg Microbes Infect, 8, 2019
|
|
5JYN
 
 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
1NIW
 
 | | Crystal structure of endothelial nitric oxide synthase peptide bound to calmodulin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nitric-oxide synthase, ... | | Authors: | Aoyagi, M, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-12-26 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for endothelial nitric oxide synthase binding to calmodulin
Embo J., 22, 2003
|
|
8UCX
 
 | |
6LAV
 
 | |
6LB0
 
 | |
8UW0
 
 | |
1NHL
 
 | | SNAP-23N Structure | | Descriptor: | Synaptosomal-associated protein 23 | | Authors: | Freedman, S.J, Song, H.K, Xu, Y, Eck, M.J. | | Deposit date: | 2002-12-19 | | Release date: | 2003-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homotetrameric Structure of the SNAP-23 N-terminal Coiled-coil Domain
J.Biol.Chem., 278, 2003
|
|
6LF3
 
 | |
6KW5
 
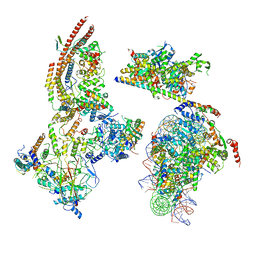 | | The ClassC RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome
To Be Published
|
|
6LJU
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-chloranyl-4-(methylamino)-2-phenyl-phenyl]amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6L22
 
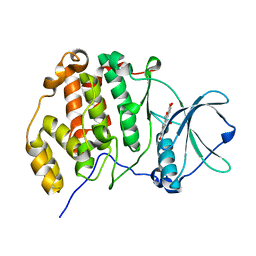 | | Crystal structure of CK2a1 H115Y with hematein | | Descriptor: | (6aR)-3,4,6a,10-tetrakis(oxidanyl)-6,7-dihydroindeno[2,1-c]chromen-9-one, Casein kinase II subunit alpha | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12318087 Å) | | Cite: | Structural insights for producing CK2 alpha 1-specific inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6L3U
 
 | | Human Cx31.3/GJC3 connexin hemichannel in the presence of calcium | | Descriptor: | CALCIUM ION, Gap junction gamma-3 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, H.J, Jeong, H, Ryu, B, Hyun, J, Woo, J.S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Cryo-EM structure of human Cx31.3/GJC3 connexin hemichannel.
Sci Adv, 6, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6LA4
 
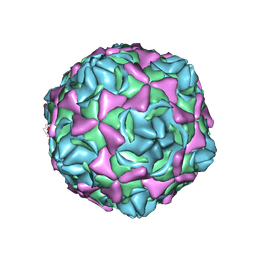 | | Cryo-EM structure of full echovirus 11 particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LAH
 
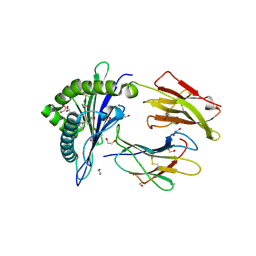 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LFG
 
 | |
6VPP
 
 | | Cryo-EM structure of microtubule-bound KLP61F motor with tail domain in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp61F, ... | | Authors: | Bodrug, T, Wilson-Kubalek, E.M, Nithianantham, S, Debs, G, Sindelar, C.V, Milligan, R, Al-Bassam, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding.
Elife, 9, 2020
|
|
5KIF
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*GP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*CP*C)-R(P*G)-D(P*GP*TP*AP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
1JTH
 
 | | Crystal structure and biophysical properties of a complex between the N-terminal region of SNAP25 and the SNARE region of syntaxin 1a | | Descriptor: | SNAP25, syntaxin 1a | | Authors: | Misura, K.M.S, Gonzalez Jr, L.C, May, A.P, Scheller, R.H, Weis, W.I. | | Deposit date: | 2001-08-21 | | Release date: | 2001-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biophysical properties of a complex between the N-terminal SNARE region of SNAP25 and syntaxin 1a.
J.Biol.Chem., 276, 2001
|
|
6VJG
 
 | | Csx3-I222 Crystal Form at 1.8 Angstrom Resolution | | Descriptor: | CRISPR-associated protein, Csx3 family | | Authors: | Brown, S, Charbonneau, A, Burman, N, Gauvin, C.C, Lawrence, C.M. | | Deposit date: | 2020-01-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA 4 .
J.Biol.Chem., 295, 2020
|
|
