8SFJ
 
 | | WT CRISPR-Cas12a with a 10bp R-loop | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*AP*CP*TP*TP*AP*TP*CP*AP*CP*TP*AP*AP*AP*AP*GP*AP*TP*CP*GP*GP*AP*AP*G)-3'), DNA (5'-D(P*CP*TP*TP*CP*CP*GP*AP*TP*CP*TP*TP*TP*TP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Strohkendl, I, Taylor, D.W. | | Deposit date: | 2023-04-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | WT CRISPR-Cas12a with a 10bp R-loop
To Be Published
|
|
8SFH
 
 | |
8SFL
 
 | |
8SFP
 
 | |
8SJC
 
 | | Crystal structure of Zn2+ bound calprotectin | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Perera, Y.R, Garcia, V, Guillen, R.M, Chazin, W.J. | | Deposit date: | 2023-04-17 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of Zn2+ bound calprotectin
To Be Published
|
|
2H2S
 
 | |
8TTH
 
 | | NorA single mutant - D307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
7CYN
 
 | | Cryo-EM structure of human TLR7 in complex with UNC93B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein unc-93 homolog B1, ... | | Authors: | Ohto, U, Ishida, H, Shimizu, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of Toll-like receptors in complex with UNC93B1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2GLX
 
 | | Crystal Structure Analysis of bacterial 1,5-AF Reductase | | Descriptor: | 1,5-anhydro-D-fructose reductase, ACETATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dambe, T.R, Scheidig, A.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of NADP(H)-Dependent 1,5-Anhydro-d-fructose Reductase from Sinorhizobium morelense at 2.2 A Resolution: Construction of a NADH-Accepting Mutant and Its Application in Rare Sugar Synthesis
Biochemistry, 45, 2006
|
|
7D7N
 
 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
7D6B
 
 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase with delta-gluconolactone | | Descriptor: | Beta-glucosidase 18, D-glucono-1,5-lactone, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
2GIV
 
 | | Human MYST histone acetyltransferase 1 | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, Probable histone acetyltransferase MYST1, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-29 | | Release date: | 2006-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Crystal Structure of Human MYST histone acetyltransferase 1 in complex with acetylcoenzyme A
To be Published
|
|
7DFM
 
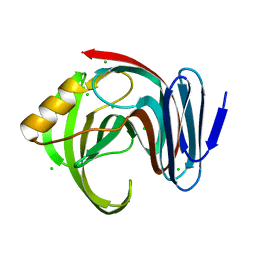 | |
7D6A
 
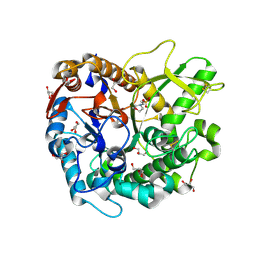 | | Crystal structure of Oryza sativa Os4BGlu18 monolignol beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 18, GLYCEROL, ... | | Authors: | Baiya, S, Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of rice Os4BGlu18 monolignol beta-glucosidase.
Plos One, 16, 2021
|
|
8TPJ
 
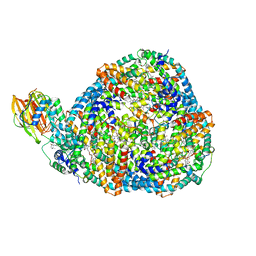 | | Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Orange carotenoid-binding protein, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-04 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TIM
 
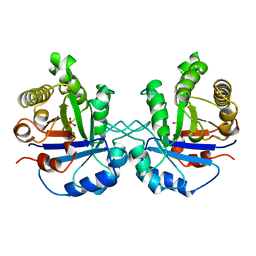 | |
2GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
7D94
 
 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound one Mg2+ and one Rb+ in the presence of bufalin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DDF
 
 | | Crystal structures of Na+,K+-ATPase in complex with beryllium fluoride | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.62 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2GPQ
 
 | |
7DFN
 
 | | Crystal structure of glycoside hydrolase family 11 beta-xylanase from Streptomyces olivaceoviridis E-86 in complex with alpha-L-arabinofuranosyl xylotetraose | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, SODIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Kaneko, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based substrate specificity analysis of GH11 xylanase from Streptomyces olivaceoviridis E-86.
Appl.Microbiol.Biotechnol., 105, 2021
|
|
7DWY
 
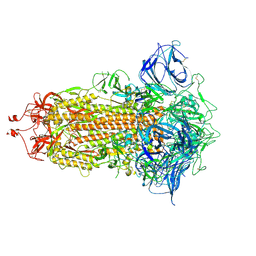 | | S protein of SARS-CoV-2 in the locked conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
8TFG
 
 | |
7DX6
 
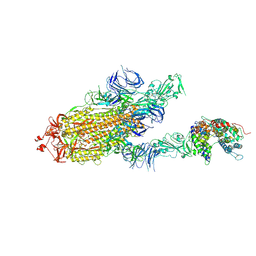 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 3 (2 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DWX
 
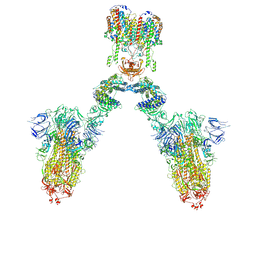 | | Conformation 1 of S-ACE2-B0AT1 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
