5ZRL
 
 | | M. smegmatis antimutator protein MutT2 in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-DIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
1XMV
 
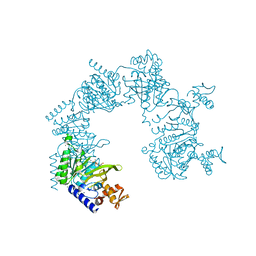 | | E. coli RecA in complex with MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RecA protein | | Authors: | Bell, C.E, Xing, X. | | Deposit date: | 2004-10-04 | | Release date: | 2005-01-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Escherichia coli RecA in Complex with MgADP and MnAMP-PNP(,).
Biochemistry, 43, 2004
|
|
5ZRC
 
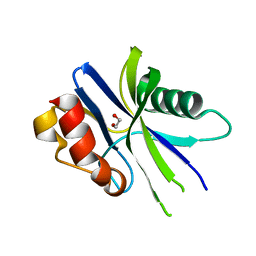 | | Structural insights into the catalysis mechanism of M. smegmatis antimutator protein MutT2 | | Descriptor: | 1,2-ETHANEDIOL, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZRH
 
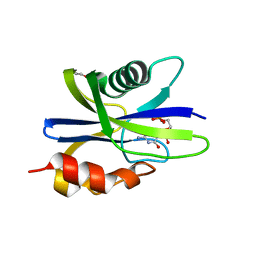 | | M. smegmatis antimutator protein MutT2 in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-MONOPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5W45
 
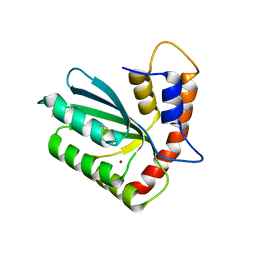 | | Crystal structure of APOBEC3H | | Descriptor: | APOBEC3H, ZINC ION | | Authors: | Ito, F, Yang, H.J, Xiao, X, Li, S.X, Wolfe, A, Zirkle, B, Arutiunian, V, Chen, X.S. | | Deposit date: | 2017-06-09 | | Release date: | 2018-03-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Understanding the Structure, Multimerization, Subcellular Localization and mC Selectivity of a Genomic Mutator and Anti-HIV Factor APOBEC3H.
Sci Rep, 8, 2018
|
|
1ZV1
 
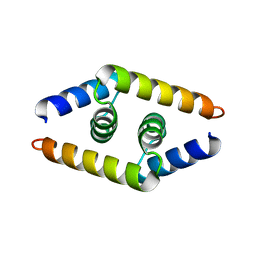 | | Crystal structure of the dimerization domain of doublesex protein from D. melanogaster | | Descriptor: | Doublesex protein | | Authors: | Weiss, M.A, Bayrer, J.R, Wan, Z, Li, B, Phillips, N.B. | | Deposit date: | 2005-06-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimerization of doublesex is mediated by a cryptic ubiquitin-associated domain fold: implications for sex-specific gene regulation
J.Biol.Chem., 280, 2005
|
|
1IZ5
 
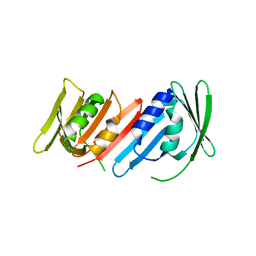 | | Pyrococcus furiosus PCNA mutant (Met73Leu, Asp143Ala, Asp147Ala): orthorhombic form | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-09-23 | | Release date: | 2003-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA
PROTEIN SCI., 12, 2003
|
|
3SWR
 
 | |
1N7A
 
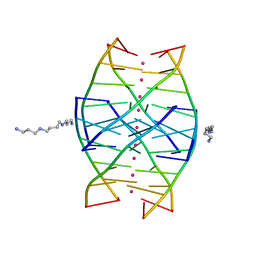 | | RIP-Radiation-damage Induced Phasing | | Descriptor: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | Authors: | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | Deposit date: | 2002-11-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
8ADK
 
 | |
8ADJ
 
 | | Poly(ADP-ribose) glycohydrolase (PARG) from Drosophila melanogaster in complex with PARG inhibitor PDD00017272 | | Descriptor: | 1-[(2,5-dimethylpyrazol-3-yl)methyl]-N-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ariza, A, Fontana, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling.
Nat Commun, 14, 2023
|
|
1AB0
 
 | | C1G/V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN AT PH 4.5 | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C, Simpson, M, Banaszak, L, Bernlohr, D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
1ACD
 
 | | V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C.D, Simpson, M, Banaszak, L.J, Bernlohr, D.A. | | Deposit date: | 1997-02-06 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
6J67
 
 | | Crystal structure of the compound 34 in a complex with TRF2 | | Descriptor: | 3FB-PHE-B8R-LEU-5XU-PRO, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, Lei, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cyclic Peptidic Mimetics of Apollo Peptides Targeting Telomeric Repeat Binding Factor 2 (TRF2) and Apollo Interaction.
ACS Med Chem Lett, 9, 2018
|
|
4BQK
 
 | | rice importin_alpha : VirD2NLS complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, IMPORTIN SUBUNIT ALPHA-1A, T-DNA BORDER ENDONUCLEASE VIRD2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-05-31 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural Basis of Interaction of Bipartite Nuclear Localization Signal from Agrobacterium Vird2 with Rice Importin-Alpha
Mol.Plant, 7, 2014
|
|
8BNY
 
 | | Structure of the tetramerization domain of pLS20 conjugation repressor Rco | | Descriptor: | CHLORIDE ION, Immunity repressor protein | | Authors: | Bernardo, N, Crespo, I, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | A tetramerization domain in prokaryotic and eukaryotic transcription regulators homologous to p53.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4NDN
 
 | | Structural insights of MAT enzymes: MATa2b complexed with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
8CIH
 
 | | Structure of FL CINP | | Descriptor: | Cyclin-dependent kinase 2-interacting protein, SODIUM ION | | Authors: | Foglizzo, M, Zeqiraj, E. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
5CZ1
 
 | |
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
1BRE
 
 | | IMMUNOGLOBULIN LIGHT CHAIN PROTEIN | | Descriptor: | BENCE-JONES KAPPA I PROTEIN BRE | | Authors: | Schormann, N, Benson, M.D. | | Deposit date: | 1995-07-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tertiary structure of an amyloid immunoglobulin light chain protein: a proposed model for amyloid fibril formation.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2WD5
 
 | | SMC hinge heterodimer (Mouse) | | Descriptor: | STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 1A, STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 3 | | Authors: | Michie, K.A, Haering, C.H, Nasmyth, K, Lowe, J. | | Deposit date: | 2009-03-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Positively Charged Channel within the Smc1/Smc3 Hinge Required for Sister Chromatid Cohesion.
Embo J., 30, 2011
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
8FOM
 
 | | Crystal structure of tRNA^Lys(SUU) bound to UAA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
5CZ2
 
 | | Crystal structure of a two-domain fragment of MMTV integrase | | Descriptor: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | Authors: | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
