2CVZ
 
 | |
1SMF
 
 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
2CU6
 
 | |
2CUT
 
 | |
1WU7
 
 | | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum | | Descriptor: | Histidyl-tRNA synthetase | | Authors: | Tanaka, Y, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum
To be Published
|
|
2CW8
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
1SN4
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
2CWY
 
 | | Crystal structure of conserved hypothetical protein, TTHA0068 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA0068 | | Authors: | Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved hypothetical protein, TTHA0068 from Thermus thermophilus HB8
To be Published
|
|
2CY5
 
 | | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-2 crystal) | | Descriptor: | CALCIUM ION, epidermal growth factor receptor pathway substrate 8-like protein 1 | | Authors: | Mizohata, E, Hamana, H, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-2 crystal)
To be Published
|
|
1WW1
 
 | |
2CZ5
 
 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3 | | Descriptor: | CITRIC ACID, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Arai, R, Ito, K, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Pyrococcus horikoshii OT3
To be Published
|
|
2CZK
 
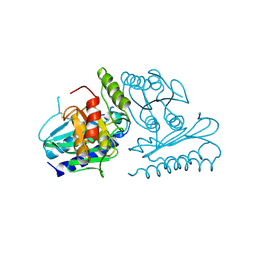 | | Crystal structure of human myo-inositol monophosphatase 2 (IMPA2) (trigonal form) | | Descriptor: | Inositol monophosphatase 2 | | Authors: | Arai, R, Ito, K, Hanawa-Suetsugu, K, Ohnishi, T, Ohba, H, Yoshikawa, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures
Proteins, 67, 2007
|
|
1WWB
 
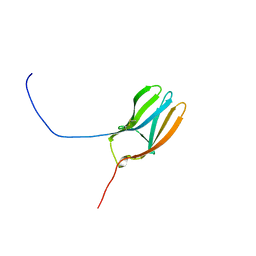 | | LIGAND BINDING DOMAIN OF HUMAN TRKB RECEPTOR | | Descriptor: | PROTEIN (Brain Derived Neurotrophic Factor Receptor TrkB) | | Authors: | Wiesmann, C, Ultsch, M.H, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-05-03 | | Release date: | 1999-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
2CZG
 
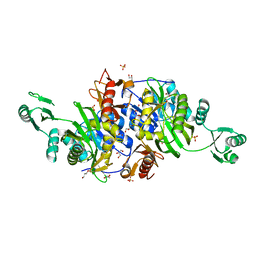 | | Crystal structure of Probable phosphoribosylglycinamide formyl transferase (PH0318) from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, SULFATE ION, phosphoribosylglycinamide formyl transferase | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Probable phosphoribosylglycinamide formyl transferase (PH0318) from Pyrococcus horikoshii OT3
To be Published
|
|
1Q8Z
 
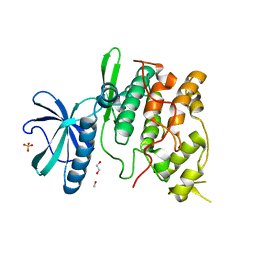 | | The apoenzyme structure of the yeast SR protein kinase, Sky1p | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, SR Protein Kinase, ... | | Authors: | Nolen, B, Ngo, J, Chakrabarti, S, Vu, D, Adams, J.A, Ghosh, G. | | Deposit date: | 2003-08-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Nucleotide-Induced Conformational Changes in the Saccharomyces cerevisiae SR Protein Kinase, Sky1p, Revealed by X-Ray Crystallography
Biochemistry, 42, 2003
|
|
1SLV
 
 | |
1SMA
 
 | | CRYSTAL STRUCTURE OF A MALTOGENIC AMYLASE | | Descriptor: | MALTOGENIC AMYLASE | | Authors: | Kim, J.S, Cha, S.S, Oh, B.H. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a maltogenic amylase provides insights into a catalytic versatility.
J.Biol.Chem., 274, 1999
|
|
1CW3
 
 | | HUMAN MITOCHONDRIAL ALDEHYDE DEHYDROGENASE COMPLEXED WITH NAD+ AND MN2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, MITOCHONDRIAL ALDEHYDE DEHYDROGENASE, ... | | Authors: | Ni, L, Zhou, J, Hurley, T.D, Weiner, H. | | Deposit date: | 1999-08-25 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Human liver mitochondrial aldehyde dehydrogenase: three-dimensional structure and the restoration of solubility and activity of chimeric forms.
Protein Sci., 8, 1999
|
|
1SQA
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-4-(PYRIMIDIN-2-YLAMINO)-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1D5R
 
 | | Crystal Structure of the PTEN Tumor Suppressor | | Descriptor: | L(+)-TARTARIC ACID, PHOSPHOINOSITIDE PHOSPHATASE PTEN | | Authors: | Lee, J.O, Yang, H, Georgescu, M.-M, Di Cristofano, A, Pavletich, N.P. | | Deposit date: | 1999-10-11 | | Release date: | 1999-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association.
Cell(Cambridge,Mass.), 99, 1999
|
|
1WJ9
 
 | | Crystal structure of a CRISPR-associated protein from thermus thermophilus | | Descriptor: | CRISPR-associated protein | | Authors: | Ebihara, A, Yao, M, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain
Protein Sci., 15, 2006
|
|
1SH9
 
 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | POL polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, McKenna, R, Abanje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-25 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
2CVC
 
 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Hildenborough) | | Descriptor: | HEME C, High-molecular-weight cytochrome c precursor | | Authors: | Suto, K, Sato, M, Shibata, N, Kitamura, M, Morimoto, Y, Takayama, Y, Ozawa, K, Akutsu, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c
To be Published
|
|
1SHF
 
 | |
2CWC
 
 | |
