4LLH
 
 | | Substrate bound outward-open state of the symporter BetP | | Descriptor: | 2-(trimethyl-lambda~5~-arsanyl)ethanol, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, CHLORIDE ION, ... | | Authors: | Perez, C, Faust, B, Ziegler, C. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na(+) coupling.
Nat Commun, 5, 2014
|
|
4F4R
 
 | | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop
to be published
|
|
4JN7
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE
To be Published
|
|
4DW8
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I | | Descriptor: | Haloacid dehalogenase-like hydrolase, SODIUM ION, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I
To be Published
|
|
4DXK
 
 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate | | Descriptor: | Mandelate racemase / muconate lactonizing enzyme family protein, PHOSPHATE ION, SODIUM ION | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue, na and phosphate
to be published
|
|
4BEM
 
 | | Crystal structure of the F-type ATP synthase c-ring from Acetobacterium woodii. | | Descriptor: | ACETATE ION, F1FO ATPASE C1 SUBUNIT, F1FO ATPASE C2 SUBUNIT, ... | | Authors: | Matthies, D, Meier, T, Yildiz, O. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Structure and Mechanism of an F/V-Hybrid Rotor Ring in a Na+-Coupled ATP Synthase
Nat.Commun., 5, 2014
|
|
4PU6
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ cations | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5K1F
 
 | | Crystal structure of a class C beta lactamase/compound2 complex | | Descriptor: | Beta-lactamase, CADMIUM ION, INOSINIC ACID | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | GMP and IMP Are Competitive Inhibitors of CMY-10, an Extended-Spectrum Class C beta-Lactamase.
Antimicrob. Agents Chemother., 61, 2017
|
|
3LWX
 
 | |
5K1D
 
 | | Crystal structure of a class C beta lactamase/compound1 complex | | Descriptor: | Beta-lactamase, CADMIUM ION, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | AN, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | GMP and IMP Are Competitive Inhibitors of CMY-10, an Extended-Spectrum Class C beta-Lactamase.
Antimicrob. Agents Chemother., 61, 2017
|
|
1YCE
 
 | | Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus | | Descriptor: | NONAN-1-OL, SODIUM ION, subunit c | | Authors: | Meier, T, Polzer, P, Diederichs, K, Welte, W, Dimroth, P. | | Deposit date: | 2004-12-22 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus.
Science, 308, 2005
|
|
3W8S
 
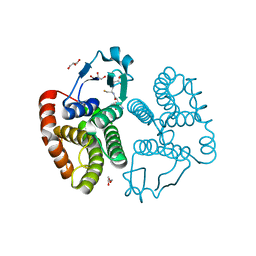 | |
2DB4
 
 | | Crystal structure of rotor ring with DCCD of the V- ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DICYCLOHEXYLUREA, SODIUM ION, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AOU
 
 | | Structure of the Na+ unbound rotor ring modified with N,N f-Dicyclohexylcarbodiimide of the Na+-transporting V-ATPase | | Descriptor: | DICYCLOHEXYLUREA, UNDECYL-MALTOSIDE, V-type sodium ATPase subunit K | | Authors: | Mizutani, K, Yamamoto, M, Yamato, I, Kakinuma, Y, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2QVK
 
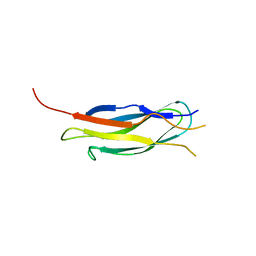 | |
3S63
 
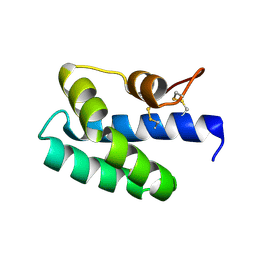 | | Saposin-like protein Na-SLP-1 | | Descriptor: | Saposin-like protein | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-24 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
1A8N
 
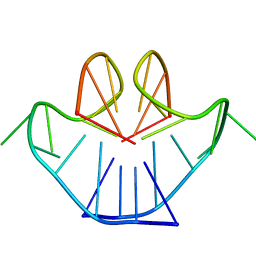 | | SOLUTION STRUCTURE OF A NA+ CATION STABILIZED DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS FORMED BY G-G-G-C REPEATS OBSERVED IN AAV AND HUMAN CHROMOSOME 19, NMR, 8 STRUCTURES | | Descriptor: | DNA QUADRUPLEX CONTAINING G.G.G.G AND G.C.G.C TETRADS | | Authors: | Kettani, A, Bouaziz, S, Gorin, A, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA.
J.Mol.Biol., 282, 1998
|
|
3A3Y
 
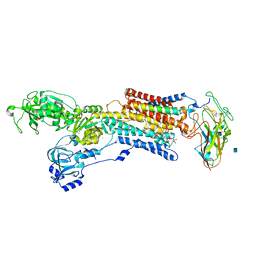 | | Crystal structure of the sodium-potassium pump with bound potassium and ouabain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Ogawa, H, Shinoda, T, Cornelius, F, Toyoshima, C. | | Deposit date: | 2009-06-23 | | Release date: | 2009-09-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sodium-potassium pump (Na+,K+-ATPase) with bound potassium and ouabain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1BKS
 
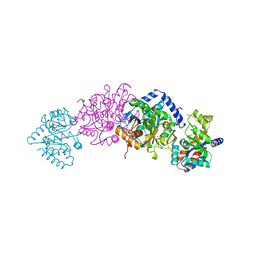 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE | | Authors: | Hyde, C.C. | | Deposit date: | 1998-07-10 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
Biochemistry, 35, 1996
|
|
2R6H
 
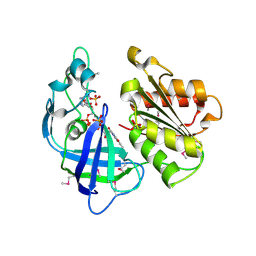 | | Crystal structure of the domain comprising the NAD binding and the FAD binding regions of the NADH:ubiquinone oxidoreductase, Na translocating, F subunit from Porphyromonas gingivalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH:ubiquinone oxidoreductase, Na translocating, ... | | Authors: | Kim, Y, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Domain Comprising the Regions Binding NAD and FAD from the NADH:Ubiquinone Oxidoreductase, Na Translocating, F Subunit from Porphyromonas gingivalis.
To be Published
|
|
2BL2
 
 | | The membrane rotor of the V-type ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, SODIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2005-02-25 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Rotor of the Vacuolar-Type Na- ATPase from Enterococcus Hirae
Science, 308, 2005
|
|
2FWU
 
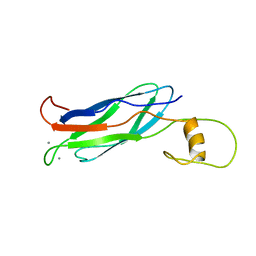 | |
1S47
 
 | | Crystal structure analysis of the DNA quadruplex d(TGGGGT)S2 | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', SODIUM ION, THALLIUM (I) ION | | Authors: | Caceres, C, Wright, G, Gouyette, C, Parkinson, G, Subirana, J.A. | | Deposit date: | 2004-01-15 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+1/Na+1 ions
Nucleic Acids Res., 32, 2004
|
|
2QVM
 
 | | The second Ca2+-binding domain of the Na+-Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Chaptal, V, Mercado Besserer, G, Abramson, J, Cascio, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The second Ca2+-binding domain of the Na+ Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2K3C
 
 | | Structural and Functional Characterization of TM IX of the NHE1 Isoform of the Na+/H+ Exchanger | | Descriptor: | TMIX peptide | | Authors: | Reddy, T, Ding, J, Li, X, Sykes, B.D, Fliegel, L, Rainey, J.K. | | Deposit date: | 2008-05-01 | | Release date: | 2008-06-03 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Transmembrane Segment IX of the NHE1 Isoform of the Na+/H+ Exchanger.
J.Biol.Chem., 283, 2008
|
|
