9AX7
 
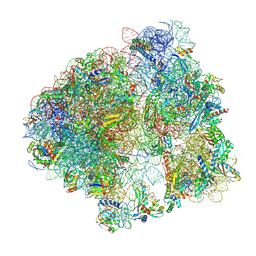 | | 70S initiation complex (tRNA-fMet M1 + CUG start codon) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mattingly, J.M, Nguyen, H.A, Dunham, C.M. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural analysis of noncanonical translation initiation complexes.
J.Biol.Chem., 300, 2024
|
|
9DR5
 
 | |
9BLN
 
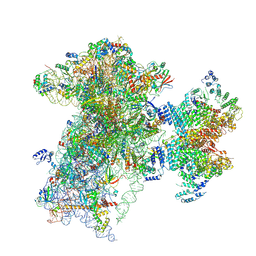 | | The structure of human Pdcd4 bound to the 40S-eIF4A-eIF3-eIF1 complex | | Descriptor: | 40S ribosomal protein S14, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Yifei, D, Albacete-Albacete, L, Ramakrishnan, V, S Fraser, C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
8Y4A
 
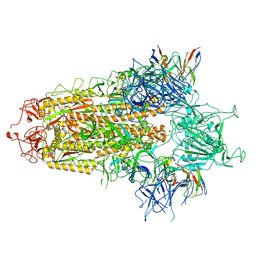 | | BA.2.86 S-trimer in complex with Nab XG2v046 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, Q, Liu, P. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Enhancing RBD exposure and S1 shedding by an extremely conserved SARS-CoV-2 NTD epitope.
Signal Transduct Target Ther, 9, 2024
|
|
9BOQ
 
 | | Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer) | | Descriptor: | 3-(2,6-difluoro-4-{[(4P)-5-{[(2S)-hexan-2-yl]sulfanyl}-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}phenyl)prop-2-yn-1-yl (1-methylpiperidin-4-yl)carbamate, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
9CGM
 
 | | The Structure of Spiroplasma Virus 4 | | Descriptor: | Capsid protein VP1, DNA binding protein ORF8 | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The Structure of Spiroplasma Virus 4 : Exploring the Capsid Diversity of the Microviridae.
Viruses, 16, 2024
|
|
9F40
 
 | | Crystal structure of the NTD domain from S. cerevisia Niemann-Pick type C protein NCR1 with ergosterol bound | | Descriptor: | ACETONITRILE, DI(HYDROXYETHYL)ETHER, ERGOSTEROL, ... | | Authors: | Nel, L, Olesen, E, Frain, K.M, Pedersen, B.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and biochemical analysis of ligand binding in yeast Niemann-Pick type C 1-related protein
To Be Published
|
|
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
9FO3
 
 | |
9EZL
 
 | |
9CWS
 
 | | Bufavirus 1 at pH 2.6 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9C6W
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (2 molecules/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9ES9
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with inhibitor DBMIB bound at plastoquinol oxidation site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9GGJ
 
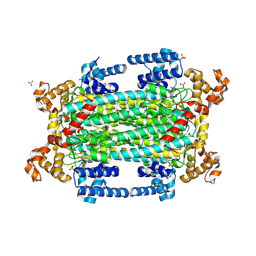 | | Crystal structure of argininosuccinate lyase from Arabidopsis thaliana (AtASL) in complex with biological substrate and products - argininosuccinate, argnine and fumarate | | Descriptor: | ARGININE, ARGININOSUCCINATE, Argininosuccinate lyase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A.J, Krzeszewska, D, Sekula, B. | | Deposit date: | 2024-08-13 | | Release date: | 2024-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Arabidopsis thaliana argininosuccinate lyase structure uncovers the role of serine as the catalytic base.
J.Struct.Biol., 216, 2024
|
|
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-22 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9GBK
 
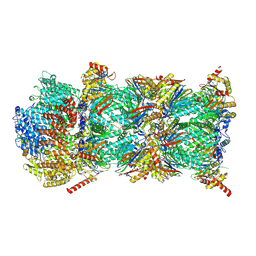 | | Blm10-20S proteasome complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome activator BLM10, Proteasome subunit alpha type-1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-07-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
9B45
 
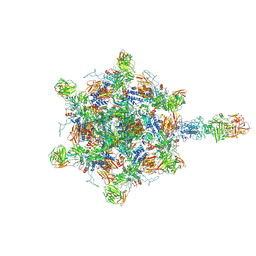 | |
9AXN
 
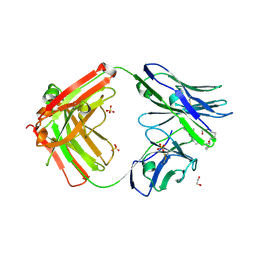 | |
9GCT
 
 | | Rho-ATP-Psu complex II expanded | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polarity suppression protein, ... | | Authors: | Gjorgjevikj, D, Wahl, M.C, Hilal, T, Loll, B. | | Deposit date: | 2024-08-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rho-ATP-Psu complex II expanded
To Be Published
|
|
9B8O
 
 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3, Vo | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-03-31 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
9JER
 
 | | Arginine decarboxylase in Aspergillus oryzae, ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-tryptophan decarboxylase PsiD-like domain-containing protein | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
8AQ6
 
 | |
5MFD
 
 | | Designed armadillo repeat protein YIIIM''6AII in complex with pD_(KR)5 | | Descriptor: | CALCIUM ION, Capsid decoration protein,pD_(KR)5, YIIIM''6AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
2LMA
 
 | | Solution structure of CD4+ T cell derived peptide Thp5 | | Descriptor: | Thp5 peptide | | Authors: | Pandey, N.K, Khan, M.M, Chatterjee, S, Dwivedi, V.P, Tousif, S, Das, J, Van Kaer, L. | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CD4+ T cell derived novel peptide Thp5 induces IL-4 production in CD4+ T cells to direct T helper 2 cell differentiation
J.Biol.Chem., 2011
|
|
6R95
 
 | |
