4JQ2
 
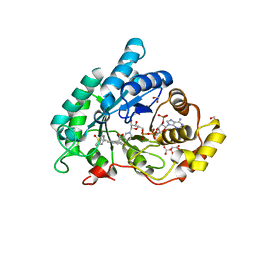 | | AKR1C2 complex with sulindac | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, L(+)-TARTARIC ACID, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of NSAID selectivity for the aldo-keto reductase 1C family
To be Published
|
|
3U9N
 
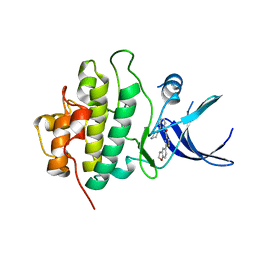 | | X-ray crystal structure of compound 1 bound to human CHK1 kinase domain | | Descriptor: | 2-(2,3-dihydro-1-benzofuran-5-yl)-N-[2-(piperazin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2011-10-19 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Novel Series of CHK1 Kinase Inhibitors with Distinctive Hinge Binding Mode
ACS MED.CHEM.LETT., 2012
|
|
4JAY
 
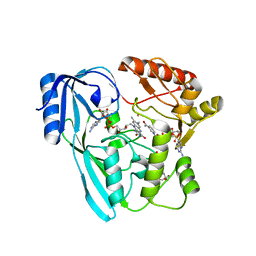 | | Crystal structure of P. aeruginosa MurB in complex with NADP+ | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chen, M.W, Lohkamp, B, Schnell, R, Lescar, J, Schneider, G. | | Deposit date: | 2013-02-19 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates.
Plos One, 8, 2013
|
|
5GQV
 
 | |
3T3S
 
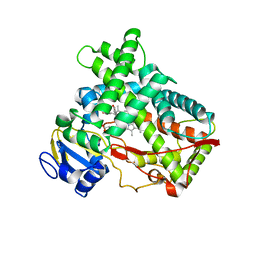 | | Human Cytochrome P450 2A13 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A13, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
4FS3
 
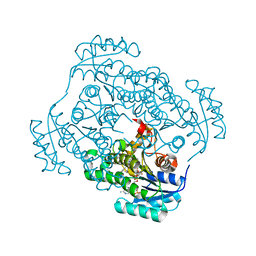 | | Crystal structure of Staphylococcus aureus enoyl-ACP reductase in complex with NADP and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH] FabI, N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, [[(2R,3S,4R,5R)-5-(3-aminocarbonyl-4H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kaplan, N, Yethon, J, Bardouniotis, E, Thalakada, R, Albert, M, Awrey, D.E, Romanov, V, Dorsey, M, Ramnauth, J, Clarke, T.E, Schmid, M.B, Berman, J, Pauls, H.W. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mode of Action, In Vitro Activity, and In Vivo Efficacy of AFN-1252, a Selective Antistaphylococcal FabI Inhibitor.
Antimicrob.Agents Chemother., 56, 2012
|
|
1SQI
 
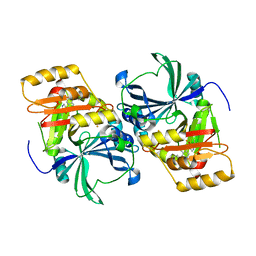 | | Structural basis for inhibitor selectivity revealed by crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvic acid dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
4I8P
 
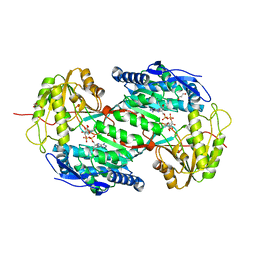 | | Crystal structure of aminoaldehyde dehydrogenase 1a from Zea mays (ZmAMADH1a) | | Descriptor: | 1,2-ETHANEDIOL, Aminoaldehyde dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
4AJO
 
 | | rat LDHA in complex with 2-((4-(4-((3-((2-methyl-1,3-benzothiazol-6yl) amino)-3-oxo-propyl)amino)-4-oxo-butyl)phenyl)methyl)propanedioic acid | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, L-LACTATE DEHYDROGENASE A CHAIN, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Watson, M, Ward, R, Tart, J, Davies, G, Caputo, A, Pearson, S. | | Deposit date: | 2012-02-16 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
4AJL
 
 | | rat LDHA in complex with 3-(ethylcarbamoylamino)-N-(2-methyl-1,3- benzothiazol-6-yl)propanamide | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, L-LACTATE DEHYDROGENASE A CHAIN, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Vogtherr, M, Ward, R, Tart, J, Davies, G, Addie, M, Ferguson, A. | | Deposit date: | 2012-02-16 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
4AL4
 
 | | rat LDHA in complex with 2-((4-(2-((3-((2-methyl-1,3-benzothiazol-6- yl)amino)3-oxo-propyl)carbamoylamino)ethoxy)phenyl)methylpropanedioic acid | | Descriptor: | 2-[[4-[2-[[3-[(2-methyl-1,3-benzothiazol-6-yl)amino]-3-oxidanylidene-propyl]carbamoylamino]ethoxy]phenyl]methyl]propanedioic acid, GLYCEROL, L-LACTATE DEHYDROGENASE A CHAIN | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Ward, R, Tart, J, Davies, G, Hohson, M, Pearson, S. | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
4AJK
 
 | | rat LDHA in complex with N-(2-(methylamino)-1,3-benzothiazol-6-yl) acetamide | | Descriptor: | GLYCEROL, L-LACTATE DEHYDROGENASE A CHAIN, MALONATE ION, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Vogtherr, M, Ward, R, Tart, J, Davies, G, Greenwood, R. | | Deposit date: | 2012-02-16 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
2CX3
 
 | | Crystal structure of a bacterioferritin comigratory protein peroxiredoxin from the Aeropyrum pernix K1 (form-1 crystal) | | Descriptor: | bacterioferritin comigratory protein | | Authors: | Mizohata, E, Murayama, K, Idaka, M, Tatsuguchi, A, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a bacterioferritin comigratory protein peroxiredoxin from the Aeropyrum pernix K1 (form-1 crystal)
To be Published
|
|
3ZGN
 
 | | Crystal structures of Escherichia coli IspH in complex with TMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-3-methyl-4-sulfanylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
2XAP
 
 | |
3FLK
 
 | | Crystal Structure of Tartrate Dehydrogenase from Pseudomonas putida in complex with NADH, oxalate and metal ion | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, AMMONIUM ION, ... | | Authors: | Malik, R, Viola, R.E. | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of tartrate dehydrogenase: a versatile enzyme catalyzing multiple reactions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1PEV
 
 | | Crystal Structure of the Actin Interacting Protein from Caenorhabditis Elegans | | Descriptor: | Actin interacting protein 1 | | Authors: | Vorobiev, S, Mohri, K, Fedorov, A.A, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-05-22 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments.
J.Biol.Chem., 279, 2004
|
|
4K9U
 
 | | Complex of human CYP3A4 with a desoxyritonavir analog | | Descriptor: | Cytochrome P450 3A4, N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-[(2S,5S)-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-D-valinamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
5XPN
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{S})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
3ZGL
 
 | | Crystal structures of Escherichia coli IspH in complex with AMBPP a potent inhibitor of the methylerythritol phosphate pathway | | Descriptor: | (2E)-4-amino-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
1CW4
 
 | |
6AG0
 
 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | Authors: | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
2AHR
 
 | | Crystal Structures of 1-Pyrroline-5-Carboxylate Reductase from Human Pathogen Streptococcus pyogenes | | Descriptor: | FORMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Nocek, B, Lezondra, L, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-28 | | Release date: | 2005-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of Delta(1)-Pyrroline-5-carboxylate Reductase from Human Pathogens Neisseria meningitides and Streptococcus pyogenes
J.Mol.Biol., 354, 2005
|
|
3WIC
 
 | | Structure of a substrate/cofactor-unbound glucose dehydrogenase | | Descriptor: | Glucose 1-dehydrogenase, PENTAETHYLENE GLYCOL, S-1,2-PROPANEDIOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
