7DW9
 
 | |
2RLT
 
 | | phosphorylated CPI-17 (22-120) | | Descriptor: | Protein phosphatase 1 regulatory subunit 14A | | Authors: | Eto, M. | | Deposit date: | 2007-08-11 | | Release date: | 2008-07-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation-induced conformational switching of CPI-17 produces a potent myosin phosphatase inhibitor.
Structure, 15, 2007
|
|
1U2B
 
 | | Triglycine variant of the Grp1 Pleckstrin Homology Domain unliganded | | Descriptor: | Cytohesin 3, SULFATE ION | | Authors: | Cronin, T.C, DiNitto, J.P, Czech, M.P, Lambright, D.G. | | Deposit date: | 2004-07-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains
Embo J., 23, 2004
|
|
2TPK
 
 | |
2RUD
 
 | |
2RUC
 
 | |
2RSC
 
 | |
2UVF
 
 | |
2VIU
 
 | | INFLUENZA VIRUS HEMAGGLUTININ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
1R6Q
 
 | | ClpNS with fragments | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-16 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
1TVR
 
 | | HIV-1 RT/9-CL TIBO | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-04-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
2VIS
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, (ESCAPE) MUTANT WITH THR 131 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
3AUQ
 
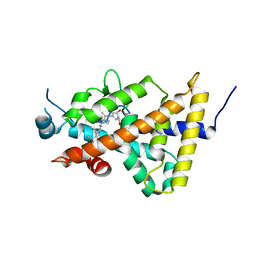 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with Yne-diene type analog of active 14-epi-2alpha-methyl-19-norvitamin D3 | | Descriptor: | (1R,2S,3R)-5-[2-[(1R,3aS,7aR)-1-[(2R)-6-hydroxy-6-methyl-heptan-2-yl]-7a-methyl-1,2,3,3a,6,7-hexahydroinden-4-yl]ethynyl]-2-methyl-cyclohex-4-ene-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Development of 14-epi-19-nortachysterol and its unprecedented binding configuration for the human vitamin D receptor
J.Am.Chem.Soc., 133, 2011
|
|
2V8K
 
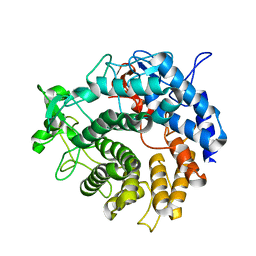 | |
2V0P
 
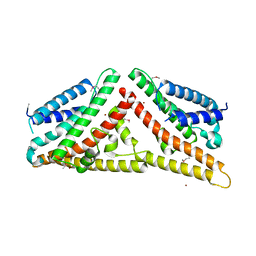 | | The Structure of Tap42 Alpha4 Subunit | | Descriptor: | TYPE 2A PHOSPHATASE-ASSOCIATED PROTEIN 42, ZINC ION | | Authors: | Roe, S.M, Yang, J, Barford, D. | | Deposit date: | 2007-05-15 | | Release date: | 2007-07-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Tap42/Alpha4 Reveals a Tetratricopeptide Repeat-Like Fold and Provides Insights Into Pp2A Regulation.
Biochemistry, 46, 2007
|
|
1R6O
 
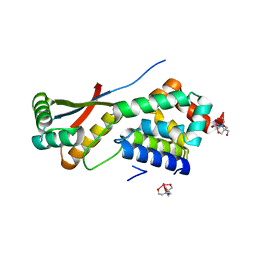 | | ATP-dependent Clp protease ATP-binding subunit clpA/ATP-dependent Clp protease adaptor protein clpS | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
2VA1
 
 | | Crystal structure of UMP kinase from Ureaplasma parvum | | Descriptor: | PHOSPHATE ION, URIDYLATE KINASE | | Authors: | Egeblad-Welin, L, Welin, M, Wang, L, Eriksson, S. | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Investigations of Ureaplasma Parvum Ump Kinase - a Potential Antibacterial Drug Target
FEBS J., 274, 2007
|
|
2WZV
 
 | | Crystal structure of the FMN-dependent nitroreductase NfnB from Mycobacterium smegmatis | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, NFNB PROTEIN, ... | | Authors: | Bellinzoni, M, Manina, G, Riccardi, G, Alzari, P.M. | | Deposit date: | 2009-12-03 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biological and Structural Characterization of the Mycobacterium Smegmatis Nitroreductase Nfnb, and its Role in Benzothiazinone Resistance
Mol.Microbiol., 77, 2010
|
|
2VIT
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, MUTANT WITH THR 155 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, LAMBDA), ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
1RJL
 
 | | Structure of the complex between OspB-CT and bactericidal Fab-H6831 | | Descriptor: | Fab H6831 H-chain, Fab H6831 L-chain, Outer surface protein B | | Authors: | Becker, M, Bunikis, J, Lade, B.D, Dunn, J.J, Barbour, A.G, Lawson, C.L. | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Investigation of Borrelia burgdorferi OspB, a BactericidalFab Target.
J.Biol.Chem., 280, 2005
|
|
1R6C
 
 | | High resolution structure of ClpN | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
1UWB
 
 | | TYR 181 CYS HIV-1 RT/8-CL TIBO | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-11-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
7EW4
 
 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW2
 
 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW3
 
 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
