1WFB
 
 | |
1WBN
 
 | | fragment based p38 inhibitors | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-(3-TERT-BUTYL-1H-PYRAZOL-5-YL)-N'-{4-CHLORO-3-[(PYRIDIN-3-YLOXY)METHYL]PHENYL}UREA | | Authors: | Cleasby, A, Devine, L.A, Gill, A.L, Jhoti, H. | | Deposit date: | 2004-11-04 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
1WBU
 
 | | Fragment based lead discovery using crystallography | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, RIBONUCLEASE | | Authors: | Cleasby, A, Hartshorn, M.J, Murray, C.W, Jhoti, H, Tickle, I.J. | | Deposit date: | 2004-11-05 | | Release date: | 2005-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1X3K
 
 | | Crystal structure of a hemoglobin component (TA-V) from Tokunagayusurika akamusi | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin component V | | Authors: | Kuwada, T, Hasegawa, T, Sato, S, Sato, I, Ishikawa, K, Takagi, T, Shishikura, F. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of two hemoglobin components from the midge larva Propsilocerus akamusi (Orthocladiinae, Diptera).
Gene, 398, 2007
|
|
1WDN
 
 | | GLUTAMINE-BINDING PROTEIN | | Descriptor: | GLUTAMINE, GLUTAMINE BINDING PROTEIN | | Authors: | Sun, Y.-J, Rose, J, Wang, B.-C, Hsiao, C.-D. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: comparisons with other amino acid binding proteins.
J.Mol.Biol., 278, 1998
|
|
2DF4
 
 | | Structure of tRNA-Dependent Amidotransferase GatCAB complexed with Mn2+ | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ammonia channel couples glutaminase with transamidase reactions in GatCAB
Science, 312, 2006
|
|
1WEJ
 
 | | IGG1 FAB FRAGMENT (OF E8 ANTIBODY) COMPLEXED WITH HORSE CYTOCHROME C AT 1.8 A RESOLUTION | | Descriptor: | CYTOCHROME C, E8 ANTIBODY, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mylvaganam, S.E, Paterson, Y, Getzoff, E.D. | | Deposit date: | 1998-03-26 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of FabE8-cytochrome c complex to 1.8 A resolution and FabE8 to 2.26 A resolution.
J.Mol.Biol., 281, 1998
|
|
1WIO
 
 | |
1WK2
 
 | |
1WMW
 
 | | Crystal structure of geranulgeranyl diphosphate synthase from Thermus thermophilus | | Descriptor: | geranylgeranyl diphosphate synthetase | | Authors: | Suto, K, Nishio, K, Nodake, Y, Hamada, K, Kawamoto, M, Nakagawa, N, Kuramitu, S, Miura, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of geranulgeranyl diphosphate synthase from Thermus thermophilus
To be Published
|
|
2BVR
 
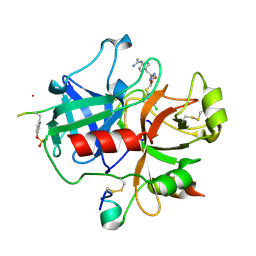 | | Human thrombin complexed with fragment-based small molecules occupying the S1 pocket | | Descriptor: | 2-[2-(4-CHLORO-PHENYLSULFANYL)-ACETYLAMINO]-3-(4-GUANIDINO-PHENYL)-PROPIONAMIDE, ALPHA THROMBIN, HIRUDIN VARIANT-2 | | Authors: | Neumann, T, Junker, H.-D, Keil, O, Burkert, K, Ottleben, H, Gamer, J, Sekul, R, Deppe, H, Feurer, A, Tomandl, D, Metz, G. | | Deposit date: | 2005-07-04 | | Release date: | 2006-10-25 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Thrombin Inhibitor Fragments from Chemical Microarray Screening
Lett.Drug Des.Discovery, 2, 2005
|
|
1WOH
 
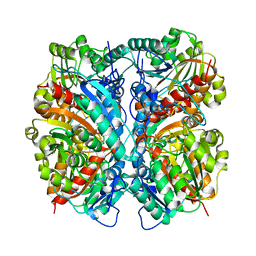 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WQ1
 
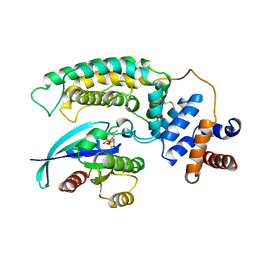 | | RAS-RASGAP COMPLEX | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, H-RAS, ... | | Authors: | Scheffzek, K, Ahmadian, M.R, Kabsch, W, Wiesmueller, L, Lautwein, A, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants.
Science, 277, 1997
|
|
1WNS
 
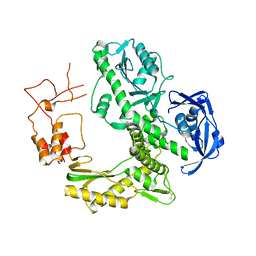 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
4I9Y
 
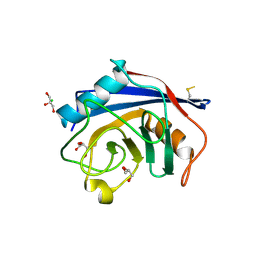 | | Structure of the C-terminal domain of Nup358 | | Descriptor: | CHLORIDE ION, E3 SUMO-protein ligase RanBP2, GLYCEROL, ... | | Authors: | Lin, D.H, Zimmermann, S, Stuwe, T, Stuwe, E, Hoelz, A. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Analysis of the C-Terminal Domain of Nup358/RanBP2.
J.Mol.Biol., 425, 2013
|
|
1WXY
 
 | |
2BU4
 
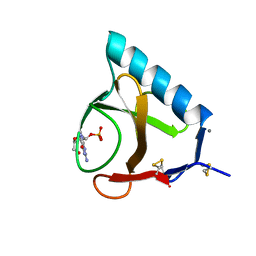 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
2BVS
 
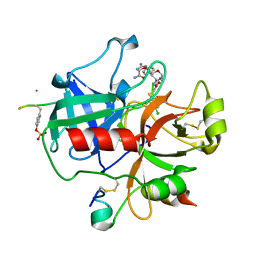 | | Human thrombin complexed with fragment-based small molecules occupying the S1 pocket | | Descriptor: | ALPHA THROMBIN, HIRUDIN VARIANT-2, N-[2-(2-CARBAMOYLMETHOXY-ETHOXY)-ETHYL]-2-[2-(4-CHLORO-PHENYLSULFANYL)-ACETYLAMINO]-3-(4-GUANIDINO-PHENYL)-PROPIONAMIDE | | Authors: | Neumann, T, Junker, H.-D, Keil, O, Burkert, K, Ottleben, H, Gamer, J, Sekul, R, Deppe, H, Feurer, A, Tomandl, D, Metz, G. | | Deposit date: | 2005-07-04 | | Release date: | 2006-10-25 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of Thrombin Inhibitor Fragments from Chemical Microarray Screening
Lett.Drug Des.Discovery, 2, 2005
|
|
2BXT
 
 | | Design and Discovery of Novel, Potent Thrombin Inhibitors with a Solubilizing Cationic P1-P2-Linker | | Descriptor: | 6-CHLORO-1-(2-{[(5-CHLORO-1-BENZOTHIEN-3-YL)METHYL]AMINO}ETHYL)-3-[(2-PYRIDIN-2-YLETHYL)AMINO]-1,4-DIHYDROPYRAZIN-2-OL, ALPHA THROMBIN, HIRUDIN VARIANT-2 | | Authors: | Bulat, S, Bosio, S, Grabowski, E, Papadopoulos, M.A, Cerezo-Galvez, S, Rosenbaum, C, Matassa, V.G, Ott, I, Metz, G, Schamberger, J, Sekul, R, Feurer, A. | | Deposit date: | 2005-07-27 | | Release date: | 2006-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and Discovery of Novel, Potent Pyrazinone-Based Thrombin Inhibitors with a Solubilizing P1-P2-Linker
Lett.Drug Des.Discovery, 3, 2006
|
|
1X0T
 
 | | Crystal structure of ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3 | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Kakuta, Y, Ishimatsu, I, Numata, T, Kimura, K, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2005-03-29 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Ribonuclease P Protein Ph1601p from Pyrococcus horikoshii OT3: An Archaeal Homologue of Human Nuclear Ribonuclease P Protein Rpp21(,)
Biochemistry, 44, 2005
|
|
1WWW
 
 | |
7DBN
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
1WX2
 
 | | Crystal Structure of the oxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase complexed with a caddie protein prepared by the addition of hydrogenperoxide | | Descriptor: | COPPER (II) ION, MelC, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
1WY2
 
 | |
1X11
 
 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
